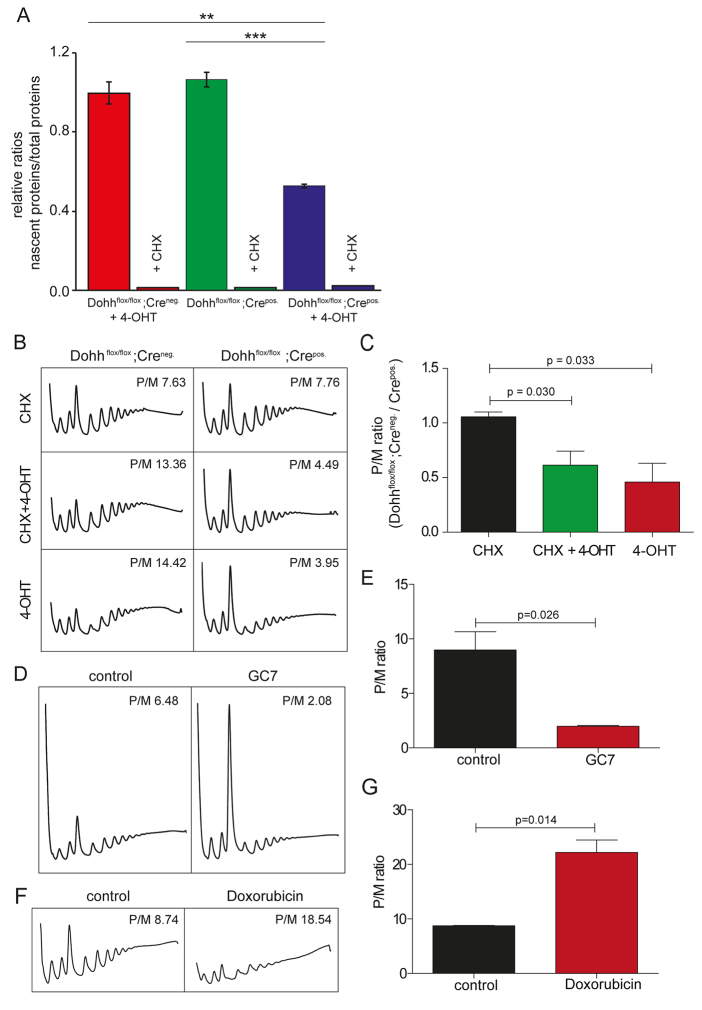
Fig. 6.
Dohh deletion impairs protein synthesis. (A) Dohh depletion causes a reduction in the rate of protein synthesis. Cells were treated for 9 days with tamoxifen or were mock-treated as indicated. Newly synthesized proteins were labelled using bioorthogonal (‘Click’) chemistry. Label incorporated into newly synthesized proteins was normalized to total protein content to determine the relative rate of protein synthesis. Note the strong reduction in label incorporation in cells treated with CHX, which demonstrates that labelling depends on new protein synthesis and indicates the large dynamic range of the assay. (B) Polysome profile comparison of 3T3 cells with tamoxifen-inducible Cre removal of DOHH (Dohhflox/flox; Crepos.) with those without (Dohhflox/flox; Creneg.). Uninduced cells (CHX) show similar polysome profiles and P/M ratios. After removal of DOHH (Dohhflox/flox; Crepos.: CHX/4-OHT and 4-OHT), profiles showed a clear increase in 80S, coupled with a mild reduction in polysomes, consistent with a translation initiation defect. (C) Ratios of P/M Crepos. to P/M Creneg. for each condition described in A. Tamoxifen-induced cells show significantly reduced ratios in comparison with uninduced cells (CHX to CHX/4-OHT, P=0.03; CHX to 4-OHT, P=0.033; n=3; one-tailed, type 3 t-test). (D) Polysome profiles of 3T3 cells treated with GC7 to inhibit DHS activity. Profiles generated by GC7 treatment phenocopy those produced by removal of Dohh, but the effect is stronger. (E) Quantification of P/M ratios shows a significant difference between control and GC7-treated 3T3 cells (P=0.026; n=3; one-tailed, type 3 t-test). (F,G) Polysome profile and P/M ratio quantification for 3T3 cells treated with doxorubicin, an inhibitor of cell growth that does not directly interfere with translation. Note the absence of a Dohh-like initiation defect in doxorubicin-treated cells. Data are displayed as mean ± s.e.m. P-values were determined using Student’s unpaired t-test (**P<0.01, ***P<0.001).