Figure 6.
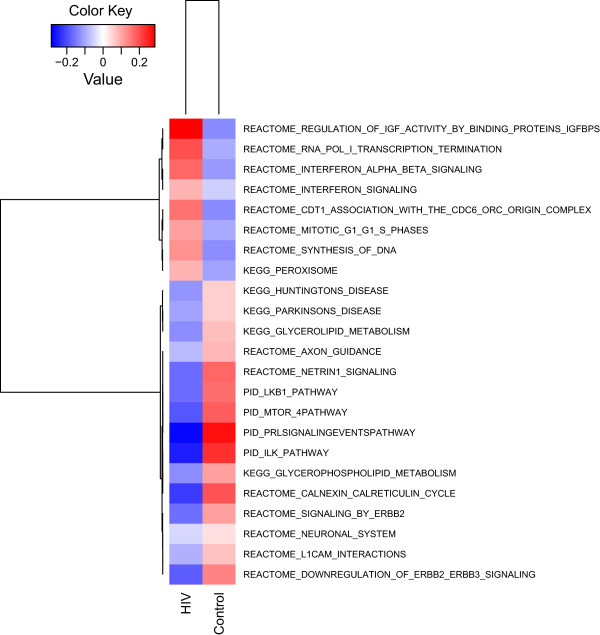
Top 23 differentially regulated pathways in the hippocampus of HIV-1 Tg rats and controls by GSEA. The Gene Set Enrichment Analysis (GSEA) algorithm was used for pathway analysis using the MSigDB C2 canonical pathway collection [28]. Of the significantly differentially regulated pathways with statistical significance of p ≤ 0.01, 8 showed increased activation, while the rest was decreased. Among the increased ones are pathways consistent with astrocyte and microglia activation, inflammatory processes, and interferon activation. Decreased pathways indicate substantial downregulation of signaling systems involved in neuronal trophism and synaptic synaptic function. Pathways are colored according to their expression values: red indicates upregulated genes in HIV-1 Tg rats, while blue indicates downregulated pathways. Brightness is proportional to the extent of change in pathway expression.
