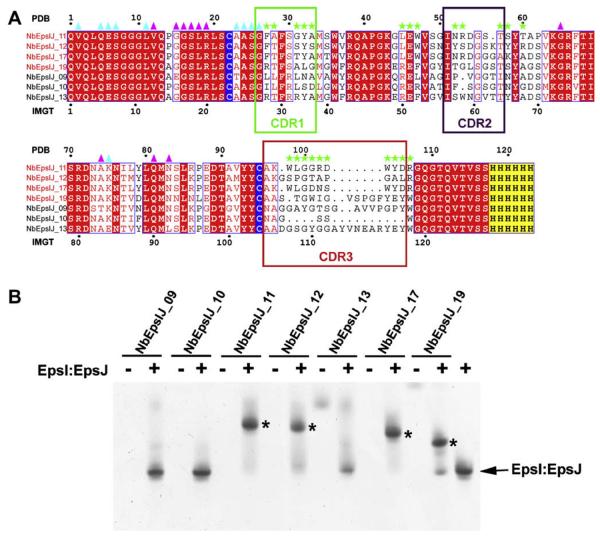
Fig. 1.
Anti-EpsI:EpsJ generated nanobodies and complex formation with EpsI:EpsJ. (A) Sequence alignment of nanobodies generated against V. vulnificus EpsI:EpsJ. The nanobodies that bound to EpsI:EpsJ according to gel-shift assays (Fig. 1B) are titled in red. While the numbering on top of the alignment corresponds to the continuous numbering present in the PDB file, the numbering on the bottom of the alignment corresponds to the standard IMGT numbering for antibodies and related proteins (Lefranc, 2005; Supplementary Figure S1). The conserved cysteines that form the intra-molecular disulfide bridge are highlighted in blue while the C-terminal hexahistidine tag is highlighted in yellow. Boxed segments of sequence compose the CDR regions with the green box as CDR1, purple box as CDR2, and red box as CDR3. Green stars denote residues that make contacts with EpsJ. Cyan and purple triangles reflect residues that make nanobody-nanobody contacts, with cyan as those of Chain C and purple of Chain F. (B) Ternary complex formation of N-terminal His6 EpsI:EpsJ with nanobodies. A native PAGE gel of anti-EpsI:EpsJ nanobodies alone and with V. vulnificus EpsI:EpsJ. The majority of the nanobodies do not enter the gel due to the high pI's that these protein exhibit (~9). Only NbEpsIJ-13 and NbEpsIJ-19, with pI = 8.0, enter the gel and are seen in the nanobody control lanes. Ternary complex formation between EpsI:EpsJ and the nanobody (NbEpsIJ-11, NbEpsIJ-12, NbEpsIJ-17, NbEpsIJ-19) cause a dramatic band shift in the gel and are denoted by asterisks. The EpsI:EpsJ control is seen in the far right lane. (For interpretation of color mentioned in this figure the reader is referred to the web version of the article.)