Abstract
Deletion of chromosome 1p35 is a common event in epithelial malignancies. We report that DEAR1 (annotated as TRIM62) is a chromosome 1p35 tumor suppressor that undergoes mutation, copy number variation and loss of expression in human tumors. Targeted disruption in the mouse recapitulates this human tumor spectrum with both Dear1−/− and Dear1+/− mice developing primarily epithelial adenocarcinomas and lymphoma with evidence of metastasis in a subset of mice. DEAR1 loss of function in the presence of TGFβ results in failure of acinar morphogenesis, upregulation of EMT markers, anoikis resistance, migration and invasion. Furthermore, DEAR1 blocks TGFβ-SMAD3 signaling resulting in decreased nuclear phosphorylated SMAD3 by binding to and promoting the ubiquitination of SMAD3, the major effector of TGFβ-induced EMT. Moreover, DEAR1 loss increases levels of SMAD3 downstream effectors, SNAI1 and SNAI2, with genetic alteration of DEAR1/SNAI2 serving as prognostic markers of overall poor survival in an 889 invasive breast cancer cohort.
Keywords: DEAR1, tumor suppressor, EMT, TGFβ, Signaling, SMAD3 ubiquitination
INTRODUCTION
Chromosome 1p35 has long been known to harbor an important tumor suppressor(s) involved in multiple tumor types including a 1.5cM major consensus interval for pancreatic cancer and a region of high frequency loss of heterozygosity in both early and late colon cancers (1, 2). LOH in chromosome 1p35–36 has also been observed in other tumors including stomach, colon and rectum, lung, breast, endometrium, testis, kidney, thyroid, and sarcomas (3). Furthermore, deletions in chromosome 1p35–36 correlate with poor survival in colon and breast cancers (4) and shorter disease-free survival in colon cancer (5). We previously identified DEAR1 as a novel gene mapping into this genomic interval and a member of the TRIM protein family intimately associated with differentiation control as well as in oncogenesis (6, 7). DEAR1 is both mutated and homozygously deleted in breast cancer and its expression was observed to be downregulated/lost in 56% of early onset breast cancers as well as in ductal carcinoma in situ (DCIS), one of the earliest pre-invasive forms of breast cancer. DEAR1 expression predicts local recurrence-free survival in early onset breast cancer, suggestive that elucidation of the function of DEAR1 could aid in stratification of breast cancers for targeted therapies (6). Importantly, introduction of DEAR1 to complement a mutation in a breast cancer cell line restored acinar morphogenesis in 3D culture while stable knockdown in immortal HMECs recapitulated the phenotype of mutant DEAR1 cells with loss of apical-basal polarity, diffuse apoptosis and failure of lumen formation, indicating that DEAR1 regulates polarity and tissue architecture. Therefore, we hypothesized that DEAR1 is a novel tumor suppressor that the loss of function of which might be critical in the loss of polarity associated with epithelial-mesenchymal transition (EMT) (6, 8).
EMT is a complex and highly specialized developmental process in which tightly joined and polarized epithelia lose apical-basal polarity and tissue architecture, and become disassociated, spindle-shaped, mesenchymal cells capable of migration (9). The preponderance of evidence also indicates that inappropriate activation of EMT in cancer results in loss of epithelial polarity and a restructuring of tissue architecture as well as a remodeling of the extracellular matrix and actin cytoskeleton driving cancer cell migration, invasion, and ultimately metastasis (9).
The most potent inducer of EMT in epithelial cancers is the cytokine transforming growth factor β (TGFβ) (9). TGFβ elicits its effects through activation of its receptor and subsequent phosphorylation of the major effector SMADs (SMAD2/SMAD3), which in complex with SMAD4, translocate to the nucleus to transcriptionally activate context dependent gene sets that repress the proliferative response or activate EMT (10). Although much is known about the downstream molecular networks that signal EMT through TGFβ, the master regulatory controls on TGFβ’s oncogenic axis are not understood as well as the underlying mechanisms governing cell polarity and tissue architecture that are lost in the initial stages of EMT (11). More importantly, elucidation of the regulatory controls on TGFβ’s function is critical for design of novel therapies targeting the oncogenic arm of the pathway (12). We therefore hypothesized that loss of function of DEAR1 could play a causal role in the initiation of EMT in malignancies associated with 1p35 LOH or deletion. Experiments described herein address this hypothesis and identify DEAR1 as a tumor suppressor intimately linked to the regulation of TGFβ-driven EMT.
RESULTS
Dear1 is a Tumor Suppressor in the Mouse
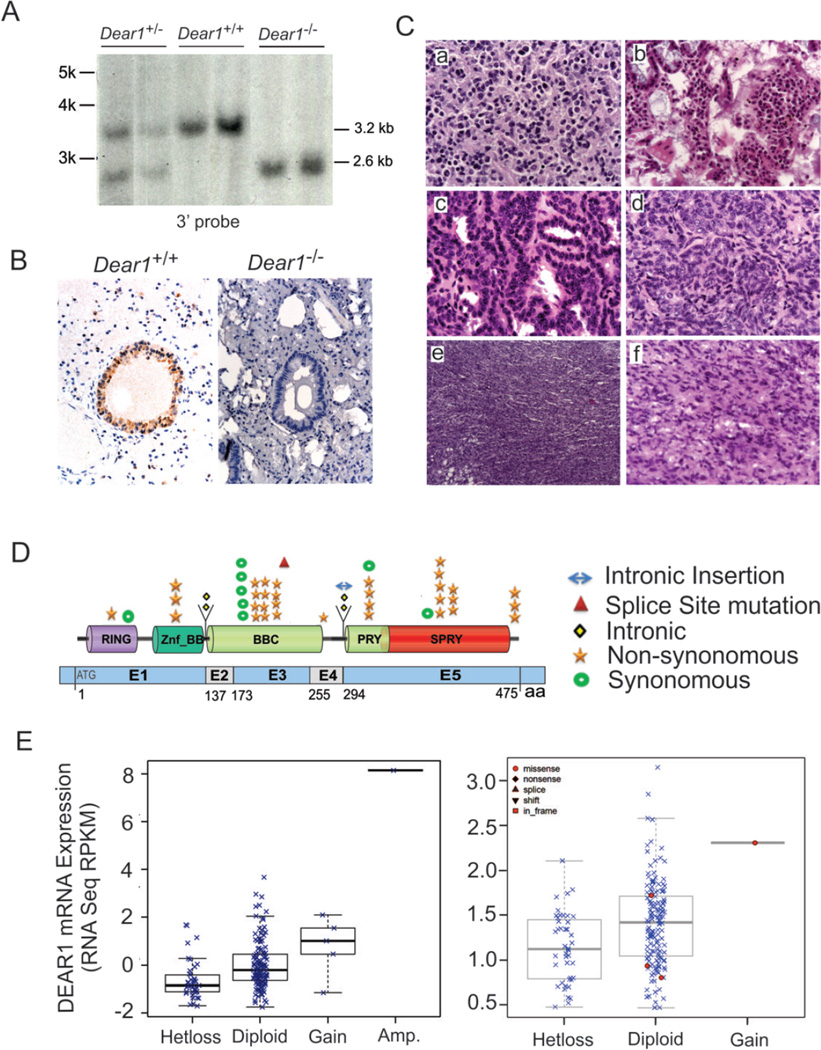
Because DEAR1 was previously found to be mutated or homozygously deleted and downregulated in breast cancer (6), we asked whether DEAR1 was a bona fide chromosome 1p35 tumor suppressor by performing targeted disruption of Dear1 in the mouse (Fig.1A; Supplementary methods and Fig. S1A). Absence of Dear1 expression was observed in Dear1−/− mouse tissues by immunohistochemistry (IHC) using an affinity purified N terminal antibody previously published (6) (Fig. 1B). RT-QPCR also confirmed the loss of DEAR1 expression in Dear+/− mice (Supplementary Fig.S1B). Genotypes performed on 30 Dear+/− x Dear1+/− mice were not significantly different from the expected Mendelian ratios (p = 0.3219) and both Dear1+/− and Dear1−/− animals were similar phenotypically to wild type littermates as well as had similar survival frequencies (averaging 20–23 months for Dear1+/− and Dear1−/− animals respectively and 21 months for wild type controls) (Supplementary Fig. S1C and D). However, Dear1−/− and Dear1+/− mice formed late onset tumors in 12.9% (8/62) and 17.7% (17/96) of animals, respectively, compared to a frequency of 4% (2/50) in wild type littermates. Significantly, the tumor spectrum of Dear1−/− and Dear1+/− mice contained adenocarcinomas from multiple organs, including mammary, pancreatic, lung and liver as well as both low and high grade sarcoma, and lymphomas on multiple organ sites, (Fig.1C and Supplementary Table S1). Mice with multiple epithelial tumors as well as metastatic adenocarcinomas were also observed (Supplementary Table S1). Thus, results indicate that Dear1 is a novel tumor suppressor and significantly, that Dear1+/− mice gave rise to tumors with a similar frequency as those observed in Dear1−/− animals, suggesting that Dear1 might function as a haploinsufficient tumor suppressor. To address this possibility representative tumor tissue sections from liver, lung, mammary gland adenocarcinomas, lymphoma and sarcoma tumors were analyzed for DEAR1 expression by IHC in Dear1+/− mice. DEAR1 expression was absent in 8/11 of Dear1+/− adenocarcinomas examined (Supplementary Fig.S1E). However, Dear1 expression was positive in 6/8 lymphoma samples, ranging from 30% positive staining cells in Hodgkins lymphoma to 80% positive staining in ureter lymphoma (Supplementary Table S2). Allele-specific PCR of Dear1+/− lymphoma samples confirmed loss of the wild type allele in the two cases of lymphoma that also showed loss of expression by IHC (Supplementary Fig.S1F). Thus, loss of the wild type allele was observed in the majority of epithelial carcinomas consistent with Dear1 behaving as a classical tumor suppressor but clearly loss was not observed in all Dear1+/− tumors, suggesting that haploinsufficiency could play a more predominant role in certain tumors, such as lymphoma. Tissue-specific haploinsufficiency has been suggested as a viable mechanism for inactivation of tumor suppressors in a context or tissue-specific manner depending on the cellular mileu in the particular tissue and in the context of other genetic hits in late onset tumors (13).
Figure 1.
(A) The knockout of Dear1 gene was identified by Southern blotting. (B) The knockout of Dear1 gene was identified in lung by immunohistochemistry. (C) H&E sections of various tumors developed in the Dear1+/− and Dear1−/− mice. (a) lymphoma in mammary gland (40x), (b) adenoma in the small intestine (40x), (c) lung adenocarcinoma (40x), (d) lung adenocarcinoma (40x), (e) sarcoma (10x), (f) sarcoma (40x). (D) Schematic representation of the multiple types of genomic alterations observed in DEAR1 domains in different tumor types. (E) DEAR1 putative copy-number alterations from GISTIC: loss of an allele of DEAR1 correlates with reduced mRNA expression in lung squamous carcinoma (left) and colorectal cancer (right).
Tumor Formation in Mouse Model Closely Recapitulates the Tumor Spectrum in Human Tumors associated with DEAR1 genetic alteration
The novel mouse model developed a variety of tumors that were strikingly similar to human tumors with chromosome 1p35 LOH. We therefore examined the human tumor spectrum for genetic alteration in DEAR1. Because DEAR1 maps into a tumor suppressor locus in pancreatic cancer (1), we performed sequencing on 55 pancreatic adenocarcinoma samples which indicated that 2/55 tumors (3.6%) contained novel missense mutations (R223H and R254Q), which were not seen in dbSNP, 1000 Genomes as well as 192 normal allele controls, and were predicted to be deleterious by PolyPhen2. In addition, we performed database analyses of TCGA, cBio (14), and ICGC (15) to determine if DEAR1 was also mutated in other chromosome 1p35-related tumors. Results in Figure 1D and Supplementary Table S3 indicate that DEAR1 undergoes rare mutation in multiple tumor types associated with chromosome 1p35 LOH including lung squamous, endometrial and renal carcinomas as well as glioblastoma multiforme and upperaerodigestive tract tumors. In colorectal carcinoma, 4 of 221 tumors contained missense mutations in DEAR1 (R190H, R223C, R307C, and D421G). Both codon 223 and codon 307 had been previously found to be mutated in pancreatic and lung adenocarcinoma, respectively, by our lab and Rudin et al 2012, correspondingly (16).
Significantly, DEAR1 also undergoes copy number alterations in a variety of different cancer cell lines and primary tumors associated with 1p35 LOH. Screening of the CONAN database indicated that copy number losses affecting the DEAR1 locus were frequent events in multiple epithelial cell lines and haematopoietic cancers (Supplementary Fig.S2A,B) (17). In addition, using the cBio database (14), we analyzed the frequency of copy number losses within the DEAR1 gene across multiple tumor types in data reported by TCGA. Results indicated that DEAR1 undergoes heterozygous loss in multiple tumor types observed in the Dear1 mouse model (Supplementary Table S4). Moreover, a putative homozygous deletion, encompassing the DEAR1 locus in glioblastoma multiforme, was discovered in the provisional TCGA cohort (14). Loss or downregulation of expression of DEAR1 was also associated with tumor types undergoing copy number losses using data from the TCGA cohorts (Fig.1E, Supplementary Table S5) (14). In addition, DEAR1 IHC was performed on a pancreatic adenocarcinoma tissue microarray which revealed a significant number of pancreatic tumor samples showing either no expression (9/32, 28%), or barely detectable levels of DEAR1 staining (11/32, 34%), indicative that 62% were downregulated for DEAR1 expression (Supplementary Fig S3A). In addition, DEAR1 expression was downregulated or undetectable in 3/4 pancreatic cancer cell lines as well as human immortalized pancreatic ductal epithelial cells (HPDE) transformed with K-Ras compared to HPDE cells that express high levels of DEAR1 (Supplementary Fig. S3B).
Our data support DEAR1’s role as a tumor suppressor based on loss/downregulation of expression, mutation and copy number loss in multiple human tumors as well as the tumor spectrum recapitulated in the Dear1 mouse model. Loss of function of Dear1 in the mouse also resulted in a subset of metastatic lesions indicative that Dear1 loss underlies not only tumorigenesis, but also metastasis. Loss of a single tumor suppressor rarely results in both primary adenocarcinomas as well as metastasis. However, it has recently been shown that well characterized tumor suppressors involved in the regulation of cell cycle and cell proliferation, such as p53 and Rb, could also function in the regulation of EMT and the initiation of metastasis (18, 19). Since our previous data indicate that DEAR1 is an important regulator of morphogenesis and cell polarity in 3D culture (6) and disruption of cell polarity is a key event associated with EMT, we further analyzed the correlation between DEAR1 mutation and clinical status. Results indicated that the majority of the human tumors with DEAR1 mutation for which clinical information was available (8/10) were invasive/metastatic (Supplementary Table S6). Based on these compelling data, we hypothesized that DEAR1 might be important in the regulation of epithelial plasticity and critical in preventing the onset of EMT.
Loss of DEAR1 Expression results in a Failure of Acinar Morphogenesis in the Presence of TGFβ
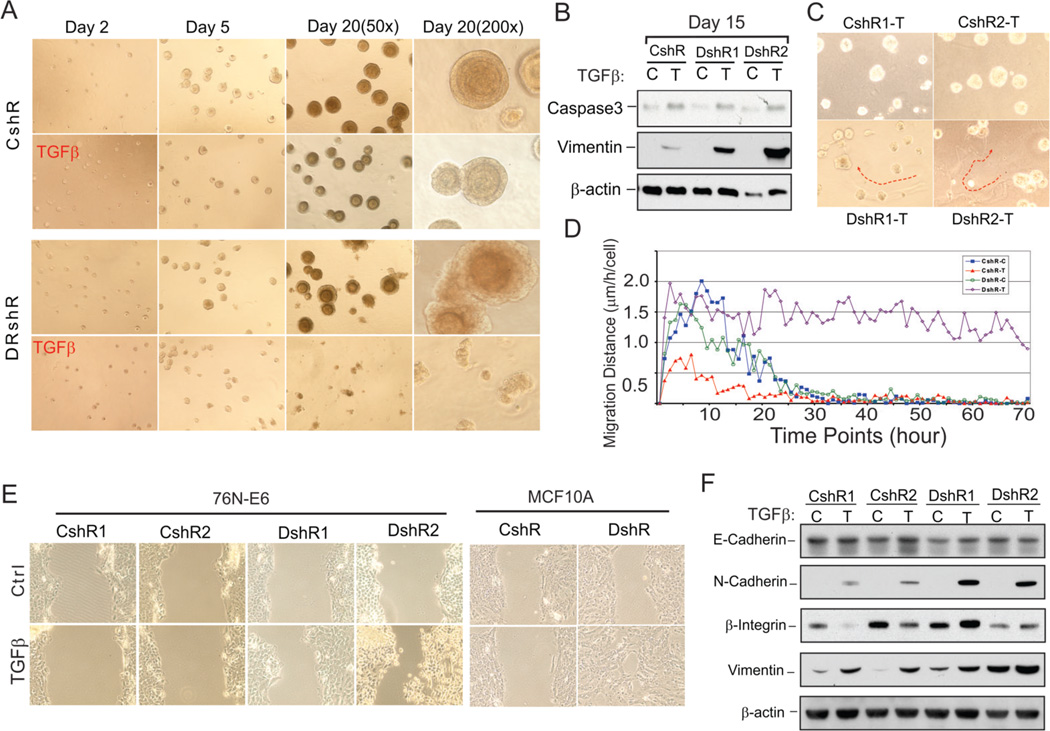
Because loss of cell polarity and tissue architecture is an important feature of epithelial cells undergoing EMT, we investigated the role of DEAR1 in the regulation of acinar morphogenesis in 3D culture in the presence or absence of TGFβ, a critical regulator of epithelial plasticity (20). Three stable DEAR1 knockdown (DEAR1-KD) clones in the immortal HMEC cell line 76N-E6 and two shRNA control clones generated previously (Supplementary Fig. S4) (6) were plated in 3D culture in the presence or absence of TGFβ. Results indicated that, in the presence or absence of TGFβ, 76N-E6 cells as well as control-shRNA clones initiated acinar morphogenesis and differentiated into uniform polar acini in the presence of TGFβ (Fig. 2A and Supplementary Fig. S5). Stable knockdown of DEAR1, however, resulted in irregularly shaped acini (Fig.2A) as we reported previously, resulting from lack of apical basal polarity and proper lumen formation (6). Strikingly, 3/3 DEAR1-KD clones propagated in 3D culture in the presence of TGFβ (2ng/ml), failed to initiate acinar morphogenesis and demonstrated a complete inability to differentiate (Fig. 2A and Supplementary figure S5). Because acini formation was not disrupted in control clones in the presence of TGFβ at same concentration, results in DEAR1-KD clones were not due to toxicity effects from the addition of TGFβ to the cultures. To determine if the failure of acinar morphogenesis in DEAR1-KD cells in the presence of TGFβ was due to apoptosis, acini collected from 3D culture were examined for caspase 3 expression. Results indicated that caspase 3 expression did not vary significantly between DEAR1-KD clones and control clones in the presence of TGFβ (Fig.2B), suggesting that failure of acini formation in DEAR1-KD clones treated with TGFβ was not a result of apoptosis. However, because TGFβ plays a critical role as a potent inducer of EMT and because mesenchymal-like cells have been reported to be unable to form acini in 3D culture (21), we examined acini for expression of Vimentin. Results indicated that Vimentin expression was significantly induced in TGFβ-treated DEAR1-KD clones compared with control clones (Fig. 2B), suggesting that in the presence of TGFβ, DEAR1-KD clones acquired mesenchymal features which prevented acinar morphogenesis. These data indicate that the presence or absence of DEAR1 dictated the formation of acini in 3D culture in the presence of the extracellular signals from TGFβ. Thus, we asked whether DEAR1 loss of function might play a causal role in the initiation of TGFβ-driven EMT.
Figure 2. DEAR1 is a negative regulator of TGFβ-induced migration and EMT.
(A) Phase contrast images of DEAR1-shRNA clone (DshR) and control-shRNA clone (CshR) with or without TGFβ (2 ng/ml) in 3D culture at indicated time. Experiments were repeated 2 times, and similar data were obtained. (B) Western analysis of DEAR1-KD clones (DshR) and control clones (CshR) in 3D culture. Cells were collected at the time points indicated with recover buffer (BD) and lysed in 1x SDS sample buffer. (C) Phase contrast images of DEAR1-shRNA clones (DshR1-T and DshR2-T) and control-shRNA clones (CshR1-T and CshR2-T) with or without (data not shown) TGFβ (2 ng/ml) in 3D culture for 5 days. Red arrows trace movement of TGFβ-treated DEAR1 shRNA clones through the matrix. (D) Cell migration distance at each time point comparing 76N-E6 DEAR1-KD clones with or without TGFβ treatment (DshR-T versus DshR-C) and control clones with or without TGFβ treatment (CshR-T versus CshR-C). The values showed are means of 150–200 cells. (E) Wound healing assay of DEAR1-KD (DshR1 and DshR2) and control clones (CshR1 and CshR2) of 76N-E6 (left panel) and MCF10A cells (DshR10A versus control shRNA (CshR10A), right panel) with or without TGFβ treatment (2ng/ml) for 24 hours. (F) Western analysis of EMT markers in 76N-E6 DEAR1-KD (DshR1 and DshR2) and control clones (CshR1 and CshR2) with or without TGFβ treatment (4 ng/ml) for 4 days.
Loss of DEAR1 Results in TGFβ-induced Cell Migration and Invasion through a 3D Matrix
Morphological changes in 2D culture were clearly evident in DEAR1-KD clones consistent with a partial mesenchymal phenotype with less compact epithelial patterning with scattering as compared with control clones which grew as compact, cobblestone-like epithelial cells (Supplementary figure. S6A). After treatment with TGFβ for 5 days, control clones showed no obvious difference in morphology, while DEAR1-KD clones showed a marked increase in scattering as well as cells displaying a prominent mesenchymal morphology (Supplementary figure. S6A). DEAR1-KD and control clones were also observed in 3D culture in the presence of TGFβ. Strikingly, prior to acinar morphogenesis, by day 5, clear traces of cell migration through the matrigel were visible in some cells from DEAR1-KD clones in the presence of TGFβ as opposed to control clones which showed no evidence of traces, indicating that the DEAR1-KD clones exhibited cell motility in the presence of TGFβ (Fig. 2C). To better quantify cell migration, we performed time lapse microscopy to observe migration following plating of DEAR1-KD and control clones with and without TGFβ in matrigel 3D cultures. Results indicated that the average distance of cell migration in both DEAR1-KD (40.28 ± 12.36 µm) and control shRNA clones (42 ± 14.7 µm) did not differ significantly in the absence of TGFβ, indicating that without TGFβ, loss of function of DEAR1 does not affect cell migration through the matrix (Fig. 2D, and Supplementary Fig. S6B, S6C). Moreover, time lapse video clearly documented that DEAR1-KD cells exhibited extensive migration on top of the matrigel and invasion through the matrigel in the presence of TGFβ (supplementary movies 1–4). The average migration distance of DEAR1 WT clones following TGFβ treatment was 16.02 ± 6.52 µm (p=0.0057 vs. no treatment), compared with a much enhanced migration distance in DEAR1-KD clones following TGFβ treatment (173.2 ± 15.19 µm, p<0.00001 vs. no treatment) (Supplementary Fig. S6C). The longest migration distance recorded for DEAR1-KD cells was 1400 µm. Like control clones, DEAR1-KD cells showed some evidence of migration without TGFβ treatment, however, the migration halted after the first 15–20 hours of experiments, while the migration in TGFβ-treated DEAR1-KD cells continued for the entire experimental period (72 hours) (Fig. 2D). Furthermore, both control clones (with or without TGFβ treatment) and DEAR1-KD clone (without TGFβ treatment) formed acini in 3D culture once their movement halted, perhaps reflecting an evolutionary adaptation of the cells due to the environmental changes (from 2D culture to 3D culture) (Fig.2A). However, the DEAR1-KD cells failed to form acini in 3D culture following TGFβ treatment (Fig. 2A and supplementary figure S5), which suggested that this type of migration might represent an EMT-related migration. Therefore, this evidence clearly indicates that loss of function of DEAR1 results in TGFβ-induced migration and invasion through the Matrigel matrix in these immortal HMEC lines. Because 3D culture more approximates growth in an in vivo environment, results also suggest the potential importance of loss of DEAR1 expression or mutation to the initiation of TGFβ-induced EMT involved in breast cancer progression.
Next, wound assays were performed to assess cell migration in 2D culture in DEAR1-KD cells versus controls. In control shRNA clones, TGFβ treatment failed to accelerate cell migration across the wound, while in DEAR1-KD clones, TGFβ treatment resulted in rapid wound closure compared with untreated cells (Fig. 2E, left panel). We then generated stable shRNA DEAR1 knockdown clones in the breast epithelial line MCF10A (DEAR1-KD-10A) and performed the same experiments. Results were consistent with experiments in 76N-E6 cells in that migration was accelerated in the presence of TGFβ in DEAR1-KD-10A cells compared to control shRNA cells (Fig. 2E right panel and supplementary figure S6D). To further confirm the specificity of DEAR1 KD, we generated additional DEAR1 shRNA stable clones in 76N-E6 cells (DEAR1 76N-E6 KD2) targeting a different region of DEAR1 that did not overlap with the original targeted region and carried out the same wound assays. Similar results were observed as in the initial shRNA clones, providing additional support for the specificity of the cellular KD for DEAR1 (Supplementary Fig. S6E). Additionally, we transiently transfected vector controls or DEAR1 expression constructs into U2OS which has no detectable DEAR1 expression (data not shown) and performed wound assays. Results indicated that in the vector control, TGFβ treatment enhanced cell motility and accelerated wound closure (Supplementary Fig. S6F). However, in U2OS cells ectopically expressing DEAR1, TGFβ treatment failed to significantly enhance migration across the wound, indicative that DEAR1 expression inhibited TGFβ-driven migration (Supplementary Fig. S6F). Thus, cumulative data from DEAR1-KD and ectopic expression experiments using both a 3D culture model and 2D migration models demonstrate that DEAR1 is a dominant inhibitor of TGFβ-induced cell migration and that loss of DEAR1 expression is requisite for TGFβ-driven migration and invasion in immortal HMECs.
DEAR1 Loss of Function Upregulates TGFβ-induced Early EMT Markers
We then examined the expression of EMT markers including β1-Integrin, a major effector of TGFβ-driven migration and EMT (22), in both 76N-E6 cells as well as MCF10A cells. Although the basal level of β1-Integrin varied between the two breast control lines, β1-Integrin was downregulated in both cell lines following TGFβ treatment. However, in DEAR1 KD clones in both 76N-E6 and MCF10A, β1-Integrin expression was upregulated in the presence of TGFβ (Fig. 2F and Supplementary Fig. S7A). Expression levels of E-Cadherin were slightly downregulated in DEAR1 KD clones compared with controls. However, both 76N-E6 and MCF10A KD clones showed increased expression of the EMT marker N-Cadherin. We also analyzed the expression of the mesenchymal marker Vimentin. Importantly, compared to the control clones, DEAR1-KD clones in 76N-E6 and MCF10A demonstrated increased basal expression of Vimentin (Fig. 2F and Supplementary Fig. S7A) which was further increased upon treatment with TGFβ. Upregulation of Vimentin was also confirmed by immunostaining which indicated that 13% of cells in control clones were positive for Vimentin staining, while up to 65–80% of cells in stable 76N-E6 DEAR1-KD clones showed strong Vimentin expression without TGFβ treatment (Supplementary Fig. S7B). In DEAR1 KD2 76N-E6 cells, EMT markers were upregulated similarly as reported for the initial shDEAR1 clones (Supplementary Fig. S7C). mRNA expression analysis of β1-Integrin and Vimentin by Q-RT-PCR confirmed Western analyses. TGFβ-target PAI-1 was dramatically increased in DEAR1-KD clones after TGFβ treatment (Supplementary Fig. S7D). Because upregulation of Vimentin and β1-Integrin have been shown to correlate with the induction of EMT as immediate early events in the EMT process in HMECs (23), our data indicate that DEAR1 loss of function results in immediate early induction of EMT in the presence of TGFβ.
Loss of DEAR1 promotes Anoikis-Resistance in Immortal HMEC cells
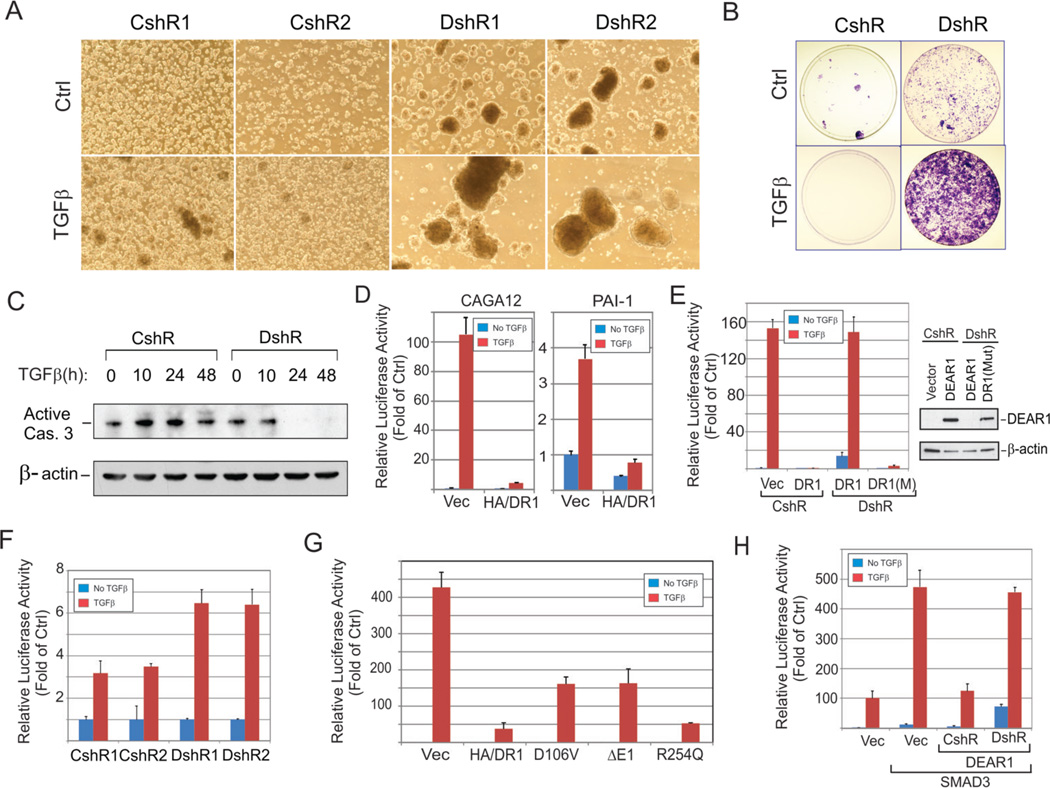
Anoikis is a process triggered by inadequate or inappropriate cell-matrix contact in which epithelial cells, separated from the epithelial layer, die by a mechanism similar to apoptosis. Resistance to anoikis confers a selective advantage to pre-cancerous epithelial cells, and also increases survival of cells detached from the matrix. Therefore, anoikis resistance is thought to be an important marker of EMT and characteristic of invasive and metastatic cells (20, 24). To examine if DEAR1 functions in the regulation of anoikis, we determined DEAR1’s ability to affect growth of cells plated on ultra-low attachment (ULA) dishes in the presence or absence of TGFβ. After 2–4 days incubation, massive cell death occurred in control 76N-E6 clones with or without TGFβ, while in DEAR1-KD clones cellular aggregates were clearly observed and greatly enhanced in the presence of TGFβ (Fig. 3A). A marked increase in colony formation was observed in DEAR1-KD clones when suspension cells were propagated in regular culture dishes, especially in the presence of TGFβ (Fig. 3B), indicative of anoikis resistance. Because anoikis is thought to be controlled in part by regulators of programmed cell death, such as caspase 3 (25), we analyzed control and KD clones for active caspase 3 expression following plating under ULA conditions without TGFβ treatment. Active caspase 3 expression was observed in control clones for the entire experimental period, up to 48 hrs, while in DEAR1-KD clones, downregulation of active caspase 3 was apparent at 24 hrs (Fig. 3C). These data indicate that DEAR1 is an important regulator of anoikis in HMEC-76NE6 cells, and that loss of function of DEAR1 drives TGFβ mediated anoikis-resistance in immortal HMECs.
Figure 3. DEAR1 loss of function is required for TGFβ-induced anoikis resistance and DEAR1 is a dominant negative regulator of TGFβ and SMAD3 signal transduction.
(A) Suspension culture of DEAR1-shRNA clones (DshR) and control clones (CshR) of 76N-E6 HMECs. Cells were seeded into ultra-low attachment (ULA) dishes with or without TGFβ treatment (2 ng/ml) for 4 days. Note: cellular aggregates in DshR1 and DshR2 cells with and without TGFβ treatment. (B) Colony formation of suspension cells. After 4-days in ULA suspension culture, cells were plated in regular tissue culture dishes, cultured for 7 days and colonies stained with crystal violet. (C) Western analysis of active caspase 3. Suspension cells were collected at the indicated time points and lysed in 1x SDS sample buffer. (D) DEAR1 inhibition of TGFβ response element-driven luciferase activity. HA-tagged DEAR1 and empty vectors were transiently co-transfected with luciferase reporters with TGFβ-response elements (CAGA12 or PAI-1) into HEK293T cells. After 24 hours, cells were treated with or without TGFβ (1 ng/ml) for 24 hours and luciferase activity measured. (E) DEAR1-mutant shRNA rescue of DEAR1 inhibition of TGFβ signaling. Along with CAGA12 luciferase reporters, empty vectors, DEAR1 (DR1) or rescue-mutant DEAR1 (DR1(mut)) were transiently co-transfected with DEAR1-shRNA (DshR) or control shRNA (CshR) into HEK293T cells. After 24 hours, cells were treated with or without TGFβ (1 ng/ml) for 24 hours and luciferase was measured. The right panel shows the Western analysis of same samples as shown in the left panel. (F) Luciferase reporter to analyze the response of DEAR1-KD clones and control clones to TGFβ (1 ng/ml). CAGA12 was transfected in cultured cells. After 24 hours, cells were treated with or without TGFβ (1 ng/ml) for 24 hours and luciferase was measured. The values were normalized with protein amount. (G) The effect of tumor-derived mutation of DEAR1 on TGFβ signal transduction. Various HA/DEAR1 mutants were co-transfected into HEK293T cells with CAGA12 reporter. After 24 hours, cells were treated with or without TGFβ (1 ng/ml) for 24 hours and luciferase was measured. (H) Co-transfection of DEAR1-shRNA with DEAR1 specifically reverses DEAR1 inhibition of SMAD3 signaling. Myc/SMAD3 was co-transfected with DEAR1 and DEAR1-shRNA or control shRNA as well as CAGA12 luciferase reporter in HEK293T cells. After 24 hours, cells were treated with or without TGFβ (1 ng/ml) for 24 hours and luciferase was measured.
DEAR1 inhibits TGFβ-induced Signal Transduction
We next investigated the role of DEAR1 in regulating TGFβ-induced signal transduction. As shown in Fig. 3D, overexpression of DEAR1 inhibited TGFβ-induced TGFβ-SMAD-response element binding site (CAGA)12 driven luciferase reporter activity (26) in HEK293T cells which have undetectable DEAR1 expression (6). Similar results were obtained with a PAI-1 luciferase reporter, a transcriptional target of TGFβ-SMAD3 signaling, in HEK293T (26). Furthermore, DEAR1 or vector control was cotransfected along with CAGA12 reporter either with DEAR1-shRNA (DshRNA) or control-shRNA (CshRNA) as well as DEAR1-rescue (DR1mut) with mutations in the shRNA-targeting region to determine if the inhibition of TGFβ signaling was specifically mediated through DEAR1. Knockdown of DEAR1 reversed the inhibitory effect of DEAR1 on the CAGA12 reporter. Furthermore, co-transfection of a mutation abolishing the shRNA targeting region in DEAR1 strongly rescued DEAR1-mediated inhibition of TGFβ signaling, indicating that the repression of TGFβ-signaling was specifically mediated by DEAR1 (Fig. 3E). TGFβ-induced signal transduction experiments were also performed in 76N-E6 DEAR1-KD cells. Results indicated that TGFβ-induced transcriptional activity was much enhanced in DEAR1-KD clones compared to control clones (Fig. 3F). We also investigated this effect of DEAR1 in the breast cancer cell line MDA-MB-231 and the cervical cancer cell line HeLa in which similar results were obtained (Supplementary Fig. S8A), indicating that introduction of DEAR1 inhibits TGFβ signaling in both breast and cervical carcinoma lines. Cumulative data indicate that DEAR1 is an important inhibitor of TGFβ-induced signal transduction. To determine if any loss of function mutations could be identified that target TGFβ signaling, DEAR1 constructs with tumor-derived missense mutations D106V and R254Q (Supplementary Table S3) were introduced with a CAGA12 reporter into HEK293T cells. Results indicated that missense mutation R254Q did not rescue TGFβ -induced luciferase activity; however, the D106V mutation resulted in partial (approximately 45%) restoration of TGFβ-induced luciferase activity compared to vector controls and similar to results obtained from deletion of the entire exon 1 (Fig. 3G). Results provide evidence that DEAR1 undergoes loss of function mutation that, in the case of the D106V mutation, restores TGFβ signaling.
DEAR1 Represses TGFβ-induced SMAD3-dependent Signaling
Because SMAD3 is a major effector of TGFβ-driven EMT (23) and both CAGA12 and PAI-1 reporters have been shown to be regulated by SMAD3 (26), we investigated whether DEAR1’s inhibition of TGFβ signaling was mediated by repression of signaling through SMAD3. Results indicated that SMAD3 transfection alone stimulated reporter luciferase activity driven by CAGA12 or PAI-1 in HEK293T and this effect was greatly enhanced after TGFβ treatment. However, co-transfection of HA-DEAR1 resulted in a dramatic inhibition of SMAD3-mediated signal transduction using either CAGA12 or PAI-1 reporters (Supplementary Fig. S8B). In addition, the inhibitory effect of DEAR1 on SMAD3 increased with increasing amounts of DEAR1 transfected, indicating a dose-dependent effect (Supplementary Fig. S8C). To confirm that the inhibitory effect was specifically mediated by DEAR1, we co-transfected DEAR1-shRNA in the system and found that DEAR1-shRNA absolutely reversed the inhibition by DEAR1 compared to the control shRNA (Fig.3H). Thus, these data demonstrate that DEAR1 inhibition of TGFβ-signal transduction is mediated, at least in part, by repression of SMAD3-dependent signaling.
DEAR1 Directly Binds to and Promotes the Ubiquitination of SMAD3
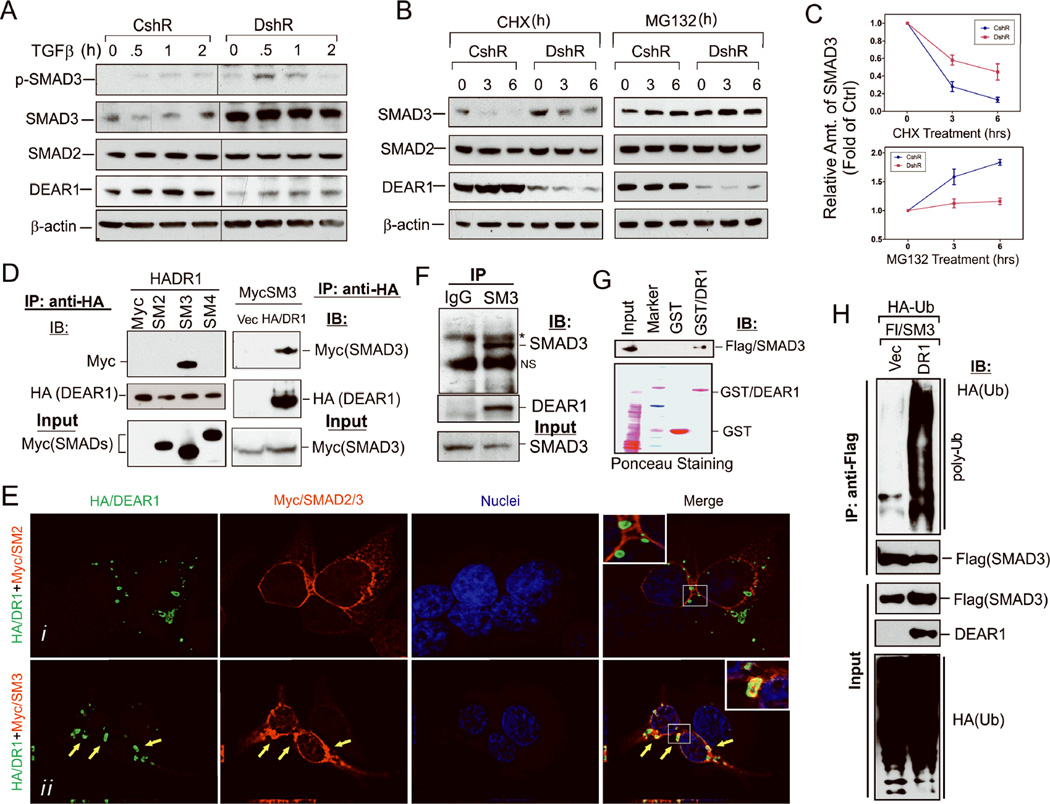
Because DEAR1 is a member of the TRIM family of proteins which have been associated with the formation and architecture of large protein complexes, we asked whether DEAR1 might bind to SMAD3 and affect SMAD3 protein stability. Endogenous SMAD protein levels were determined in 76N-E6 cells by Western analysis which indicated that SMAD2 levels did not vary between control and KD clones (Fig. 5A), whereas DEAR1-KD clones showed a dramatic increase (2–3 fold of control clones) in SMAD3 protein levels prior to TGFβ treatment and that level was not elevated in the presence of TGFβ (Fig. 5A), suggesting that DEAR1 attenuates SMAD3 protein levels independently of TGFβ treatment. Similar results were also observed using DEAR1 76N-E6 KD2 cells as well as confirmed in MCF10A DEAR1 KD cells which expressed increased SMAD3 levels relative to shRNA controls (Supplementary Fig. S9A and Supplementary Fig. S11A). To determine the mechanism underlying the increase in SMAD3 expression following DEAR1-KD, 76N-E6 DEAR1-KD and control shRNA clones were treated with the translational inhibitor cycloheximide (CHX). SMAD3 protein levels strongly decreased within 3 hours in the control cells, while in DEAR1-KD cells, SMAD3 levels are only slightly changed compared with untreated controls until 6 hrs post treatment (Fig. 4B and 4C), suggesting that DEAR1 wild type cells show reduced SMAD3 stability compared to DEAR1-KD clones. Moreover, SMAD3 levels were stabilized following treatment with the 26S proteasome inhibitor MG132 in control clones in which SMAD3 levels accumulated within 3 hrs following treatment with MG132, while there was almost no effect in DEAR1 KD in the presence of MG132, suggesting that DEAR1 mediates proteasomal degradation of SMAD3 (Fig. 4B and 4C).
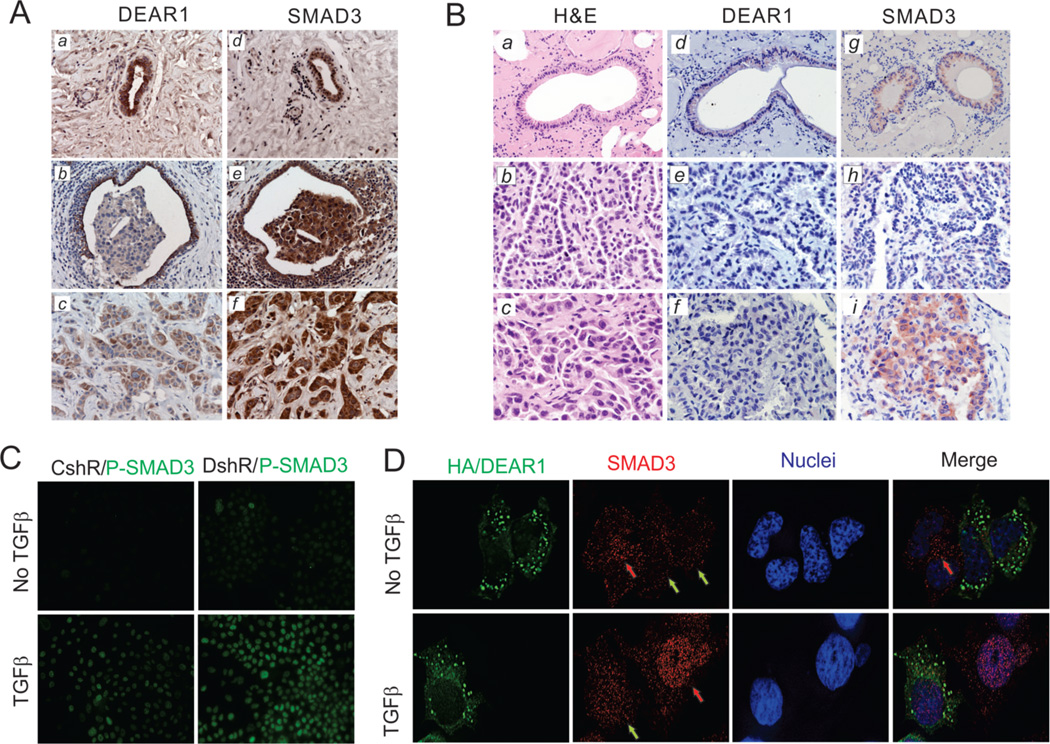
Figure 5. (A) DEAR1 and SMAD3 IHC staining in human breast cancer tissues.
DEAR1 and SMAD3 IHC staining in the normal ducts (a,d), DCIS (b,e) and carcinoma (negative staining) invading a normal duct (positive staining) (c,f) invasive carcinoma from the same individual and located in the same histologic section. Original magnification, 200x. Strong SMAD3 staining was observed in invasive carcinoma (e–f), whereas weak DEAR1 staining was observed in invasive carcinoma (b–c). (B) DEAR1 and SMAD3 IHC staining in mouse tumors. Consecutive H&E sections of normal lung (a) well-moderately differentiated lung adenocarcinoma (b) and poorly differentiated lung adenocarcinoma with high grade atypia and invasive morphology (c) from the same DEAR1+/− mouse. DEAR1 IHC staining (d–f), and SMAD3 (g–i) IHC staining in the same sections. Original magnification: a,d and g, 200x; remaining, 400x. Strong SMAD3 staining was observed in the poorly differentiated aggressive foci (i) compared to DEAR1 which showed negative staining (c). (C) DEAR1-KD increased the nuclear accumulation of phospho-SMAD3 in immortal HMECs. Low magnification deconvolution confocal images of immunofluorescence staining of phospho-SMAD3 in DEAR1-shRNA 76N-E6 cells. DEAR1-shRNA clone (DshR) and control clone (CshR) were plated on glass coverslips in 24-well plates. After 16 hours in culture, cells were serum-starved with 1:10 diluted D-Medium for 24 hrs. After treatment with or without 2 ng/ml TGFβ for 1 hour, cells growing on coverslips were fixed and immunostained with anti-phophor-SMAD3 (Millipore). All images were photographed using the same exposure conditions for comparison of images (see also Figure S10A). (D) DEAR1 transient transfection decreased endogenous SMAD3 protein expression and nuclear localization in MCF7 cells. HA/DEAR1 plasmids were transiently transfected in MCF7 cells growing on glass coverslips. After overnight incubation, cells were starved in DMEM/F12 media with 0.2% BSA for 24 hrs, then, treated with or without TGFβ (2ng/ml) for 1 hour. Cells on coverslips were fixed and co-immunostained with mouse anti-HA and rabbit anti-SMAD3. The red arrows showed SMAD3 signal in non-transfected cells (no DEAR1) and green arrows showed SMAD3 signal in DEAR1-transfected cells. Each image is representative from 5–7 fields and all images were photographed using the same exposure condition for comparison (see also Figure S10B).
Figure 4. DEAR1 binds to SMAD3 and induces SMAD3 ubiquitination.
(A) Western analysis of endogenous phospho-SMAD3, SMAD3, SMAD2 and DEAR1 expression in DEAR1-knockdown (DshR) and control (CshR) 76N-E6 HMECs. Cells were treated with TGFβ (2ng/ml) for different time points. Note dramatic increase in SMAD3 expression in DEAR1 knockdown cells. (B) Western analysis of endogenous SMAD3, SMAD2 and DEAR1 expression in DEAR1-knockdown (DshR) and control (CshR) 76N-E6 HMECs. Cells were treated with 100 ug/ml cycloheximide (CHX) or 25 uM MG132 for indicated time points. (C) Relative density analysis of Westerns (B) showing SMAD3 expression in DEAR1 knockdown versus controls at various time points following either CHX or MG132 treatment. (D) Co-immunoprecipitation (Co-IP) of HA/DEAR1 and Myc/SMAD proteins. HA/DEAR1 (HADR1) was transiently cotransfected with Myc/vector (Myc), Myc/SMAD2 (SM2), Myc/SMAD3 (SM3) or Myc/SMAD4 (SM4) into HEK293T cells shown in the left panel and Myc/SMAD3 was transiently cotransfected with HA/vector or HA/DEAR1 shown in the right panel. After 24 hours, cells were lysed in M-Per buffer (Pierce) and the lysates were immunoprecipitated with rabbit anti-HA antibody. SMAD proteins were blotted with mouse anti-Myc antibody. (E) High magnification deconvolution confocal images of immunofluorescence staining of HA/DEAR1 and Myc/SMAD2 and Myc/SMAD3. HA/DEAR1 was cotransfected with Myc/SMAD2 (panel i) or Myc/SMAD3 (panel ii) into HEK293T cells. Yellow arrows showed colocalization of DEAR1 and SMAD3. (F) Co-IP of endogenous SMAD3 and DEAR1 in HeLa cells which express both DEAR1 and SMAD3 proteins. Cell lysate was precipitated with either control anti-IgG or anti-SMAD3 (SM3) antibody. * indicates a band corresponding to the IgG heavy chain and (NS) indicates a non-specific band. (G) GST pull down assay indicates that DEAR1 directly binds SMAD3. GST-DEAR1 (GST/DR1) or control GST was added to HEK293T cell lysate expressing Flag/SMAD3 and GST was pulled down with anti-GST beads. SMAD3 was probed with a Flag antibody. (H) DEAR1 induces SMAD3 ubiquitination. DEAR1 (DR1) or empty vector (Vec) was transiently transfected with HA/Ubiquitin (HA-Ub) and Flag/SMAD3 (Fl/SM3) into HEK293T cells. Lysates were immunoprecipitated with anti-Flag antibody and probed for ubiquitination with an HA-antibody.
To investigate the role of DEAR1 in mediating the proteasomal degradation of SMAD3, we first determined if DEAR1 binds SMAD3. Results in Fig 4D show that DEAR1 co-immunoprecipitated with SMAD3, but not SMAD2 or SMAD4. We next performed immunostaining following co-transfection of HA/DEAR1 with Myc/SMAD2 or Myc/SMAD3. Results in Fig. 4E clearly show strong co-localization of DEAR1 with SMAD3 (yellow arrows, in Fig. 4Eii), while colocalization to SMAD2 was not observed (Fig. 4Ei). Furthermore, we mapped the domain of SMAD3 required to interact with DEAR1. By both co-immunoprecipitation and co-localization experiments using immunofluorescence staining, the only domains of SMAD3 involved in interacting with DEAR1 were the linker and MH2 domains of SMAD3 (Supplementary figures S9B and S9C). The endogenous interaction between SMAD3 and DEAR1 was confirmed by anti-SMAD3 pull down in HeLa cells (Fig. 4F). GST pull down assays were then performed to determine if DEAR1 directly interacts with SMAD3. GST or GST-DEAR1 fusion proteins were incubated with cell lysate from HEK293T containing transiently transfected Flag-SMAD3. Specific interaction of GST-DEAR1, but not GST alone, with Flag-SMAD3 was observed (Fig. 4G), indicating that DEAR1 physically interacts with SMAD3. Since ubiquitination of SMAD proteins has been found to be an important tool in regulation of protein stability (27), we investigated whether DEAR1 promotes ubiquitination of SMAD3 by co-transfecting Flag-SMAD3, HA-Ubiquitin and DEAR1 into HEK293T cells. SMAD3 was then pulled down by anti-Flag and ubiquitination was detected using anti-HA-tag antibodies. Results indicated that no ubiquitination was observed in vector controls, while, as indicated in Fig. 4H, DEAR1 markedly increased poly-ubiquitination of SMAD3, indicative that DEAR1 promotes the ubiquitination of SMAD3.
DEAR1 and SMAD3 Expression are Inversely Correlated in Both Human Tumors and Dear1 KO Mouse Model
To determine whether the inverse relationship between loss of function of DEAR1 and increased stability of SMAD3 observed in in vitro experiments could be recapitulated in human tumors showing loss of expression of DEAR1, we examined DEAR1 and SMAD3 expression in early onset human breast tumors compared with adjacent normal tissues. Results were striking in that in 5/7 tumor/normal pairs demonstrated an inverse correlation between DEAR1 expression and SMAD3 expression (supplementary figure Fig. S9D), while SMAD2 expression did not show an inverse relationship with DEAR1 expression. In addition, DEAR1 and SMAD3 expression was determined by IHC in 10 human DCIS with adjacent normal ductal epithelium as well as invasive carcinoma. Results showed that 4/10 demonstrated an inverse correlation in expression (Fig. 5A). In one case, a uniform staining pattern for both DEAR1 and SMAD3 was observed throughout the tumor in which DEAR1 staining was downregulated or completely absent in DCIS lesions and adjacent invasive disease contrasted with SMAD3 expression which was upregulated in DCIS and invasive breast cancers. In the remainder of cases (Supplementary Fig. S10a–f and S11a–f), tumor heterogeneity was observed with the identical region within the tumor showing loss/downregulation of DEAR1 expression correlating with upregulation of SMAD3 expression relative to staining in adjacent normal epithelium. Interestingly, the 4 cases demonstrating inverse expression were all derived from early-onset cases (ages ranging from 37–50), consistent with Western data derived from early onset tumor and adjacent normal samples (supplementary Fig. S9D). These data are also consistent with our previous finding of DEAR1 mutation as well as loss of expression correlating with local recurrence in early onset breast cancers (6). Furthermore, to confirm that the in vivo human expression studies were also recapitulated in the Dear1 mouse model, we examined the correlation between Dear1 and Smad3 expression in a heterozygous Dear1 mouse with lung adenocarcinoma for which normal bronchial epithelium, well differentiated adenocarcinoma as well as foci of poorly differentiated adenocarcinoma with high grade atypia and invasive pattern were present in the same section (Fig. 5B). Dear1 expression was detected as pale cytoplasmic staining in the normal bronchial epithelium as would be expected in a heterozygous genotype but not expressed in either the lung adenocarcinoma or associated aggressive foci (Fig.5B (d–f)). However, Smad3 expression was detected in the normal bronchial epithelium as well as in the well differentiated tumor, but strikingly upregulated in foci that were morphologically poorly differentiated with characteristics of an aggressive, invasive tumor (Fig.5B(g–i)). Cumulative data provide compelling evidence that by detailed in vitro mechanistic studies as well as by expression analyses in vivo, DEAR1 is an important regulator of SMAD3 and that loss of DEAR1 results in upregulation of SMAD3, and unbridles TGFβ -mediated EMT.
DEAR1 Knockdown Results in Increased Nuclear Accumulation of Phosphorylated SMAD3 in the Presence of TGFβ
Nuclear translocation of phosphorylated SMAD3 (p-SMAD3) after TGFβ stimulation is requisite for SMAD3’s transcriptional activation function. Since ubiquitination of SMAD3 by DEAR1 results in lower levels of SMAD3 as well as p-SMAD3 in control clones (Fig 4A), we examined localization of active p-SMAD3 by immunostaining which indicated that nuclear p-SMAD3 staining was slightly elevated in DEAR1-KD cells even without TGFβ compared with control cells, but TGFβ treatment resulted in a much increased accumulation of p-SMAD3 in the nucleus of KD cells (Fig. 5C and Supplementary Fig. S12A), indicating that DEAR1 loss of function increases the amount of p-SMAD3 in the nucleus and therefore available to transcriptionally activate EMT related genes.
To examine whether introduction of DEAR1 could attenuate SMAD3 levels in breast cancer cells, we performed transient transfection of HA/DEAR1 into MCF7 cells, which has no detectable DEAR1 protein (6). Transient transfection allowed the visualization of cells with and without DEAR1 expression along with localization and expression of SMAD3. Results in Fig. 5D demonstrate that, in the top panel (without TGFβ treatment), endogenous, primarily cytoplasmic SMAD3 levels are lower in DEAR1-over-expressing cells (green arrows) than in adjacent untransfected cells (red arrow, without DEAR1 expression), indicating that DEAR1 expression indeed degrades SMAD3 and significantly, this regulation of SMAD3 levels by DEAR1 is independent of TGFβ. In the lower panel (with TGFβ treatment), the majority of SMAD3 moved into nuclei in DEAR1-untransfected cells (red arrow), while in DEAR1-transfected cells, SMAD3 staining was less apparent and observed throughout the cell (green arrow). These data further confirm our previous observation that DEAR1 promotes the degradation of SMAD3 and thus results in less SMAD3 entering into nuclei after TGFβ stimulation in MCF7 cells. Because signal intensity of DEAR1 transfected cells precluded visualization of SMAD3 binding, we further analyzed the colocalization of DEAR1 and SMAD3 using the Imaris image analysis system which allowed three dimensional reconstruction of stacked images to determine co-localization of the two proteins (28). Using Imaris, greater than 60% of individual HA/DEAR1 signals colocalized with SMAD3 (Supplementary Fig. S12B,C). Thus, cumulative data demonstrate that DEAR1 binds to SMAD3 independently of TGFβ and regulates SMAD3 protein levels by inducing ubquitination of SMAD3, thereby DEAR1 controls the amount of SMAD3 available for phosphorylation and nuclear translocation by TGFβ.
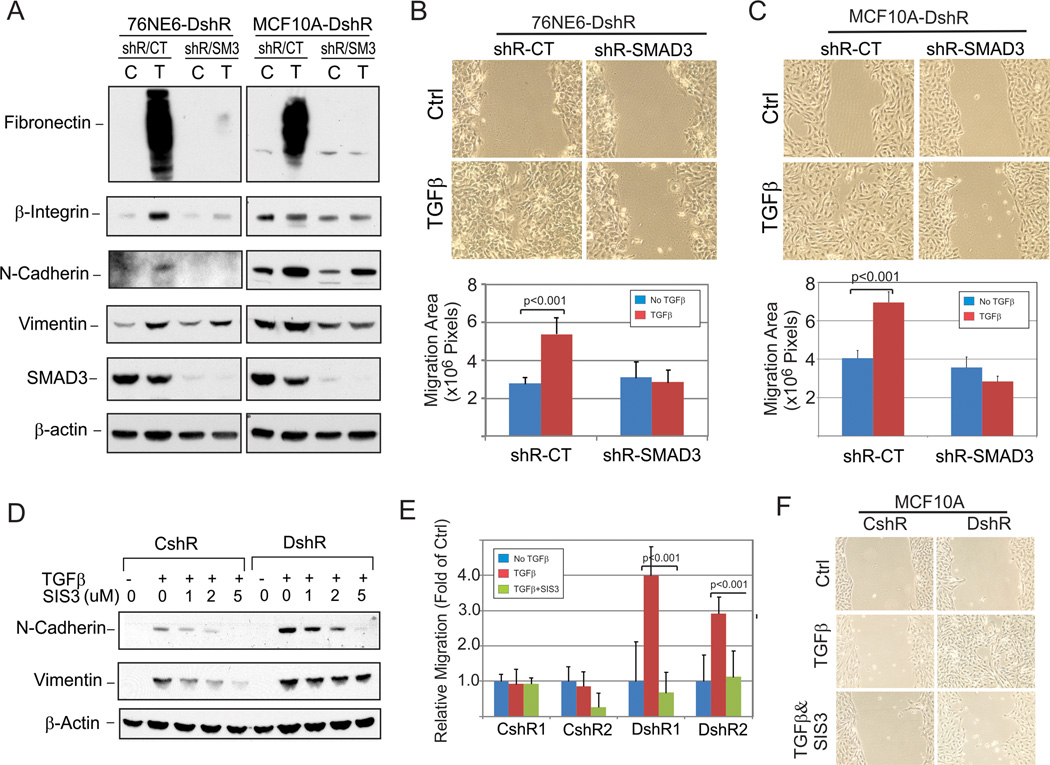
SMAD3 Loss of Function Rescues DEAR1 Loss of Function to block TGFβ-driven EMT in HMECs
To confirm that DEAR1 mediates TGFβ-induced EMT through degradation of SMAD3, we asked whether SMAD3 loss of function would rescue DEAR1 loss of function to prevent TGFβ-mediated EMT. First, we performed SMAD3 KD using SMAD3-shRNA in DEAR1-KD clones of both 76N-E6 and MCF10A lines and demonstrate that in both DEAR1 KD 76NE6DshR cells as well as MCF10A-DshR cells, SMAD3-KD strongly inhibited TGFβ-induced EMT markers (Fig. 6A) as well as greatly retarded TGFβ-induced cell migration in wound assays (Fig. 6B,C). These data clearly indicate that SMAD3-KD in DEAR1-KD cells rescued DEAR1’s loss of function phenotype. We next investigated the effect of a newly designed SMAD3 inhibitor, SIS3 (29), on TGFβ -induced EMT in DEAR1 KD clones. Results in Fig. 6D demonstrate that with increasing concentrations of SIS3, N-Cadherin and Vimentin expression induced by TGFβ were markedly attenuated. Similar results were obtained in MCF10A-DEAR1-KD cells (Supplementary Fig. S13A). We also examined the role of SIS3 in blocking TGFβ -induced cell migration in wound assays. Results in Fig. 6E show that, after TGFβ treatment, cell migration across the wound increased almost 3–4 fold in DEAR1 KD clones compared to control clones. Remarkably, the addition of SIS3 absolutely abolished TGFβ-driven cell migration in DEAR1 KD HMECs (p<0.001) as well as in DEAR1 KD MCF10A cells (Fig. 6E,F; Supplementary Fig. S13B). These data provide strong evidence that degradation of SMAD3 by DEAR1 is the primary mechanism by which DEAR1 inhibits TGFβ-induced EMT, and also give novel insight into the possibility of designing clinical inhibitors to interfere with the tumor promotion axis of the TGFβ/SMAD3 pathway.
Figure 6. (A) SMAD3-knockdown rescues TGFβ-induced EMT marker expression in DEAR1-KD 76N-E6 and MCF10A cells.
DEAR1-shRNA clones from 76NE6 and MCF10A were retrovirally infected with SMAD3-shRNA-GFP. Cells were sorted by flow cytometry to enrich knockdown cells. Then cells were plated in 6-well plates with indicated SIS3 concentrations. After 4-hour incubation, cells were treated with or without TGFβ (4ng/ml) for 3 days. Cells were dissolved in 1x SDS sample buffer and protein amount was measured using the Bradford assay (Pierce). Equal protein amount was loaded in each well for Western. (B) Wound assays demonstrate potential utility of SMAD3 knockdown to block TGFβ-induced migration across the wound in DEAR1-KD 76N-E6 cells. DEAR1-shRNA HMEC cells with SMAD3-KD (shR-SMAD3) or control-GFP (shR-CT) were plated in 6-well plates. Cells were scratched at 80% confluence and then, treated with or without 2ng/ml TGFβ for 24 hours. Phase contrast image were shown in top panel, and migration distances were quantified from 6–7 areas for each treatment (bottom panel). (C) Wound assays demonstrate potential utility of SMAD3 knockdown to block TGFβ-induced migration across the wound in DEAR1-KD MCF10A cells. DEAR1-shRNA MCF10A cells with SMAD3-KD (shR-SMAD3) or control-GFP (shR-CT) were plated in 6-well plates. Cells were scratched at 80% confluence and then, treated with or without 2ng/ml TGFβ for 24 hours. Phase contrast image were shown in top panel, and migration distances were quantified from 6–7 areas for each treatment (bottom panel). (D) Western analysis of SMAD3 inhibitor, SIS3, on TGFβ-induced EMT markers in DEAR1-KD and control clones. DEAR1-shRNA HMEC 76N-E6 clone (DshR) and control clone (CshR) were plated in 6-well plates with indicated SIS3 concentrations. After 4-hour incubation, cells were treated with or without TGFβ (4ng/ml) for 3 days. Cells were dissolved in 1x SDS sample buffer and protein amount was measured using the Bradford assay (Pierce). Equal protein amount was loaded in each well for Western. (E) Wound assays demonstrate potential utility of SMAD3 inhibitor SIS3 to block TGFβ-induced migration across the wound in DEAR1 knockdown cells. DEAR1-shRNA HMEC 76N-E6 clones (DshR1 and DshR2) and control clones (CshR1 and CshR2) were plated in 6-well plates. Cells were scratched at 80% confluence and then, treated with 2ng/ml TGFβ or TGFβ plus SMAD3 inhibitor, SIS3, for 24 hours. Migration distances were quantified from 6–7 areas for each treatment. (F) A wound assay in DEAR1-shRNA MCF10A cells treated with TGFβ and SMAD3 inhibitor (SIS3). DEAR1-shRNA clones (DshR) and control clones (CshR) were plated in 6-well plates. Cells were scratched at 90% confluence and then, treated with 2ng/ml TGFβ or TGFβ plus SMAD3 inhibitor, SIS3, for 24 hours.
Loss of DEAR1 Upregulates SMAD3 Targets SNAIL1 and SNAIL2, Master Transcriptional Regulators of EMT
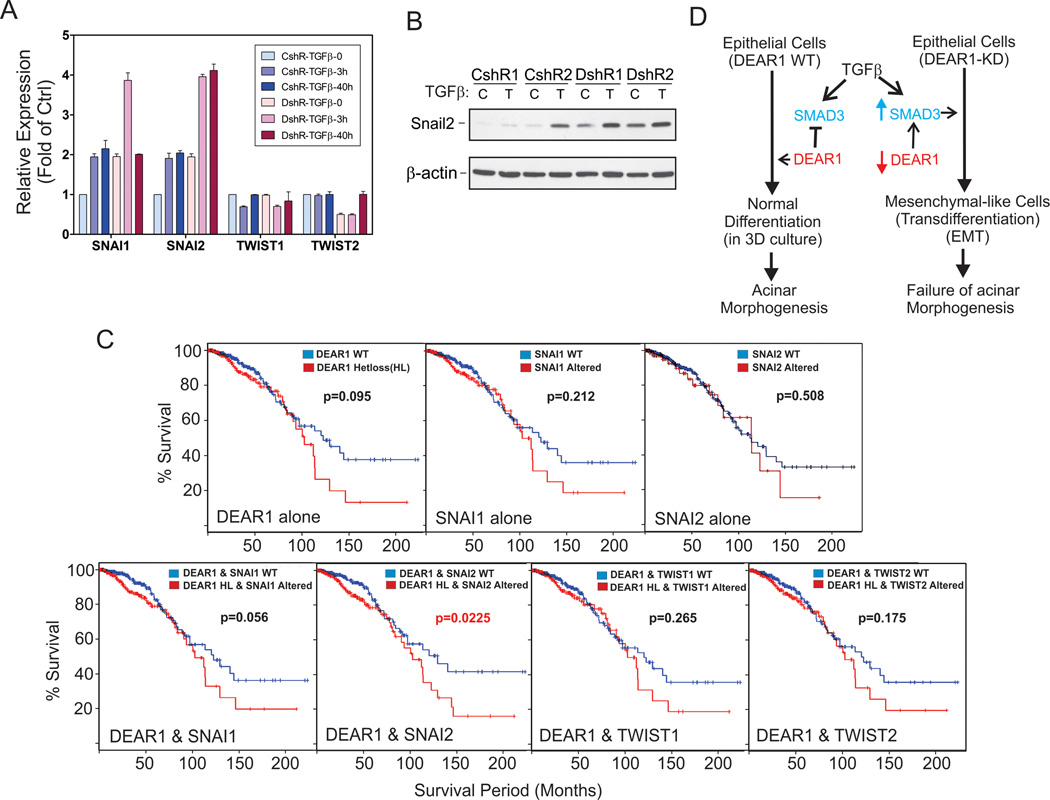
Although multiple signaling pathways initiate EMT, these pathways utilize common sets of downstream effectors as master transcriptional regulators of EMT and include SNAIL1/2, ZEB1/2 and TWIST1/2 (30, 31). Because SNAIL1 (SNAI1) and SNAIL2 (SNAI2) have been shown to be targets of TGFβ-SMAD3 signaling (32, 33), we examined mRNA levels of SNAI1/2 as well as TWIST1/2 to determine if these master transcription factors were deregulated with loss of function of DEAR1. Q-RT-PCR results indicated that the basal level of SNAI1 and SNAI2 expression increased (2 fold) compared to control clones in the 76N-E6 DEAR1 KD clone even without TGFβ treatment while TGFβ stimulation resulted in higher induction of SNAI1/2 expression (4 fold) (Fig. 7A), while no difference in expression of TWIST1/2 was observed following DEAR1 KD in the presence or absence of TGFβ (Fig. 7A). Upregulation of SNAI2 was confirmed at the protein level by Westerns which demonstrated an increase in both basal levels as well as TGFβ-inducible levels of SNAI2 (Fig. 7B). These data indicate that DEAR1 blocks a major consensus pathway downstream of TGFβ and loss of function of DEAR1 selectively upregulates SNAI but not TWIST. Because SNA1 and SNAI2 expression have been reported to be directly regulated by TGFβ-SMAD3 (32, 33), our data provide additional evidence that DEAR1’s functional role in degrading SMAD3 is a major mechanism to block TGFβ-induced EMT. To determine the clinical significance of the relationship between DEAR1 and SNAI1/2 in invasive breast cancer, we analyzed the relationship between DEAR1 loss of function including copy number variation in DEAR1, increased mRNA expression and increased protein levels, and amplification of either SNAI1 or SNAI2. Heterozygous loss of DEAR1 occurred in 33% of cases in this cohort and typically involved regions larger than the DEAR1 locus, and in some cases involved the whole short arm. Results indicated that in 889 cases of invasive breast adenocarcinoma from the TCGA cohort, accessed via cBio (14), heterozygous loss of DEAR1 trended toward significance in this large cohort (p=0.095) but by itself was not significant as a clinical indicator of poor prognosis. Likewise, SNAI1 or SNAI2 alterations alone did not significantly affect survival (p values of 0.212 and 0.508, respectively). The combination of DEAR1 heterozygous loss with SNAI1 alteration was nearly significant in predicting poor overall survival (p=0.056). However, strikingly, the combination of DEAR1 heterozygous loss with SNAI2 alteration significantly shortened overall survival (p=0.023). Furthermore, in accordance with the qPCR data, TWIST1 and TWIST2 do not significantly associate with DEAR1 heterozygous loss (p values of 0.265 and 0.175, respectively) (Fig. 7C). The data strongly support our conclusion that DEAR1 loss of function results in increased levels of SMAD3 leading to an increased expression of its downstream effectors, SNAI1 and SNAI2, and that genetic alteration of this pathway has a significant effect on patient survival.
Figure 7. (A) Quantitative RT-PCR analysis of SNAI1/2 in DEAR1-shRNA knockdown clones.
76NE6 DEAR1-KD cells were treated with or without TGFβ (4ng/ml) for 3h and 40h, then, were collected in TRIzol. RNA were extracted following TRIzol instruction and purified with Absolutely RNA microprep kit (Stratagene). Q-PCR was performed using TaqMan (Applied Biosystems). (B) Western analysis of Snail2 (Slug). 76N-E6 DEAR1-KD (DshR1 and DshR2) and control clones (CshR1 and CshR2) were treated with or without TGFβ (4 ng/ml) for 1 day. (C) The effect of DEAR1 heterozygous loss and SNAI1/2 gene upregulation on survival of invasive breast cancer patients. Survival curves were generated by cBio, using Kaplan-Meier analysis through querying complete tumor sets in the BRCA cohort for DEAR1 heterozgyous loss, SNAI1, SNAI2, TWIST1 and TWIST2. Alteration of SNAI1/2 and TWIST1/2 includes amplification, upregulation of mRNA/protein expression (if applicable) greater than two standard deviations from the mean. (D) A novel model for the regulation of TGFβ-induced EMT by DEAR1
DISCUSSION
Identification of the genetic drivers of tumorigenesis and metastasis are essential for the design of targeted therapies aimed at the underlying pathways regulated by these genes. Our combined data from the Dear1 mouse model, compilation of human tumor data and mechanistic model are consistent with DEAR1 being a critical tumor suppressor involved in many different cancers. Results presented herein from the Dear1 knockout model are striking in that loss of Dear1 in the 129/C57/BL6 background resulted in late onset tumors with a tumor spectrum similar to human tumors with LOH at chromosome 1p35 and consisting of diverse epithelial adenocarcinomas, lymphoma and sarcoma. Epithelial tumors are rarely observed in mouse models, yet, malignant epithelial adenocarcinomas are frequent in this model. Thus, these data indicate the potential importance of loss of DEAR1 to the initiation or progression of these tumor types. Because DEAR1 expression was absent in the majority of tumors analyzed, we conclude that Dear1 appears to behave as a classical tumor suppressor. However, a detailed investigation of its loss of function is warranted to determine in which circumstances haploinsufficiency might drive tumorigenesis (34, 35). Retention of the wild type allele has been observed in tumors derived from the p53 mouse models as well as other important tumor suppressors, such as pten. Interestingly, haploinsufficiency in a subset of tumors has been observed in p53 models in which the frequency of retention of the wild type p53 allele increases with increasing age of mice in which animals with tumor incidence >18 months had 85% of tumors retaining the wild type allele (35). Loss of the wild type allele could also occur very late in tumor development and could be a rate-limiting step in tumor progression (36). Thus, the novel Dear1 mouse model provides a striking example of a classical tumor suppressor that could also behave as a haploinsufficient tumor suppressor in certain tissues or contexts (35). Because chromosome 1p cytogenetic deletion is one of the most frequent deletions observed in human tumors, and because important genes involved in human tumorigenesis map distal to DEAR1 within chromosome 1p36, including CHD5 and p73 tumor suppressors (37, 38), evolution of epithelial tumors may involve not only an interstitial deletion of important loci within chromosome 1p, but also loss of the entire short arm, in which case loss of DEAR1 expression could also be pivotal to the initiation or progression of these tumor types to invasive and metastatic disease. In that regard, heterozygous loss of Dear1 in conjunction with oncogenic activation has been shown to drive tumorigenesis with dramatic differences in survival compared to controls or oncogene-activated tumors (Quintas-Cardama, et al, manuscript in preparation)
DEAR1 Loss of Function Unbridles TGFβ-induced EMT
When activated in cancer, EMT is thought to drive invasion and metastasis in breast cancer and other epithelial cancers and potentiate the generation of cells with stem-cell like characteristics resistant to current chemotherapies (39). Results in 3D culture of DEAR1-KD HMECs grown in the presence of TGFβ are consistent with an induction of a mesenchymal transdifferentiation program associated with DEAR1 downregulation resulting in an inability of these mesenchymal cells to form acinar structures in 3D culture (21). Our data also indicate that loss of DEAR1 in immortalized HMECs results in a partial EMT morphology with upregulation of Vimentin and N-Cadherin, well-known markers of early EMT induction. DEAR1 is also downregulated in a majority of DCIS lesions as well as invasive carcinoma immediately adjacent to normal ductal structures which highly expressed DEAR1 (6). Herein we demonstrate as well that DEAR1 undergoes a reduction in copy number and mRNA expression in the TCGA cohorts and that this loss of copy number is a predictor (in combination with alteration in SNAI1/2) of overall poor survival in invasive breast cancer. Thus, DEAR1 loss of function could play an early role in the initiation of EMT and could be an important biomarker of aggressive disease. In addition, cumulative results in both 2D and 3D culture indicate that DEAR1 is an important negative regulator of TGFβ-SMAD3 signaling and that loss of function in the presence of TGFβ, results in the activation of anoikis resistance, migration and invasion, key characteristics of EMT and essential steps in tumor progression and the metastatic cascade.
Ubiquitination of SMAD3 by DEAR1 restrains TGFβ-driven EMT by limiting the amount of SMAD3 protein available for phosphorylation by TGFβ
Our results also document that the interaction of DEAR1 with SMAD3 is an underlying mechanism by which DEAR1 regulates TGFβ/SMAD3-induced EMT. Although SMAD3 is highly homologous to SMAD2, recent reports found that SMAD2 and SMAD3 can play different roles in mediating TGFβ signaling. Our data indicate that DEAR1 interacted with the linker and MH2 domains of SMAD3, but did not seem to significantly affect SMAD2 which is consistent with reports that AKT binds to the linker and MH2 domains of SMAD3 but does not interact with SMAD2 (40, 41). Because DEAR1 binds to SMAD3 and induces SMAD3 ubiquitination, the rapid turnover of non-activated SMAD3 by DEAR1 may reflect the necessity for cells to more stringently control SMAD3 levels and thus the activity of this protein (42). TGFβ-induced EMT has also been shown to be associated to SMAD3-dependent and independent signal pathways (43). However, studies using SMAD3 knockout mice have indicated that TGFβ signaling through the SMAD3-dependent pathway is required for EMT (44). SMAD3 induces Vimentin transcription required for the intermediate filament system of cytoskeleton to change to a Vimentin-based cytoskeleton during EMT (23). Our data in both 2D and 3D culture indicate that loss of DEAR1 results in dramatic upregulation of Vimentin, β1-Integrin and N-Cadherin in the presence of exogenous TGFβ. These studies, together with our findings that both a SMAD3 inhibitor and SMAD3 knockdown blocked TGFβ-induced cell migration, suggest that SMAD3 is a critical mediator of TGFβ-induced EMT and that DEAR1 targets SMAD3 for degradation. Therefore, regulation of SMAD3 protein levels is a critical step in preventing EMT.
DEAR1 as a Master Regulator of the TGFB-SMAD EMT Axis
TGFβ/SMAD pathways have been shown to play roles in both tumor suppression and EMT that have always been explained as context dependent outcomes in human tumors (10). Thus, although the TGFβ/SMADs is a major tumor suppressor pathway in pancreatic cancer development, accumulating data have shown that this pathway also underlies EMT in pancreatic cancer as well as many other epithelial carcinomas (9–11). We have clearly now identified the first mechanism of action for DEAR1 as a novel and important negative regulator of TGFβ/SMAD-driven EMT. Future studies will determine whether DEAR1 plays a role in the regulation of SMAD3’s tumor suppressor functions in response to TGFβ or is a critical selective switch that dictates the context-dependent fate of TGFβ/SMAD. Future studies will also elucidate DEAR1’s role in the regulation of other key tumor suppressor pathways that could underlie its broader role in tumor suppression and unravel how DEAR1 loss of function could result in such a wide spectrum of epithelial adenocarcinomas as well as lymphoma and sarcoma.
In summary, our combined data provide critical and compelling evidence that DEAR1 as a chromosome 1p35 tumor suppressor involved in multiple human cancers as well as provide a novel paradigm for regulation of both SMAD3 protein levels and TGFβ-induced EMT. Overall, we propose a model for DEAR1 function in the presence of TGFB (Fig. 7D). In the presence of wild type DEAR1, SMAD3 is degraded and TGFβ’s oncogenic EMT arm is restrained, consistent with epithelial cells undergoing differentiation and acinar morphognesis in 3D culture. Loss of function of DEAR1 results in elevated SMAD3 available for phosphorylation and nuclear translation induced by TGFβ to activate EMT, thus, consistent with epithelial cells undergoing transdifferentiation to mesenchymal cells and a failure in acinar morphogenesis (Fig. 7D). Because TGFβ inhibitors are being used currently in clinical trials with partial success (45), our data suggest that DEAR1 could be a novel and important biomarker to stratify those breast cancer patients, and potentially other tumors demonstrating DEAR1 mutation/deletion or loss of function, for more effective treatment with specific targeted therapies aimed at the SMAD3 pathway. In addition, there is a critical need to identify novel prognostic biomarkers of aggressive disease for which increased surveillance and treatment might be necessary. Our results suggest the combined clinical utility of DEAR1 and SMAD3/SNAI2 as prognostic biomarkers for aggressive and invasive disease.
MATERIALS AND METHODS
Cell Culture and Reagents
Immortalized HMEC cells, 76N-E6, were propagated in D-Medium (6); HEK293T, U2OS and HeLa cell lines (ATCC) were propagated in DMEM with 10% FBS. MCF-7 cells were grown in DMEM/F12 medium with 10% FBS. MCF10A was cultured in medium as previously described (46). TGFβ-1 and MG132 were from Calbiochem Co., and Cycloheximide from Sigma. Luciferase kit was from Promega. (See Supplementary Methods)
Plasmids and Antibodie
DEAR1-shRNA (989-CGCCAAAGCGCTTCGATGT) and control-shRNA constructs for transient transfection were generated by inserting the synthesized oligonucleotides (Invitrogen) into the pSuper-shRNA vector which was a generous gift of Dr. Bryan R. Cullen (47). pcDNA3Myc/SMAD2, pcDNA3Myc/SMAD3 and pcDNA3Myc/SMAD4 vectors are gifts from Dr. Michelle Barton (48). HA-Ubiquitin vector was kindly provided by Dr. Edward Yeh. pcDNA3/SMAD3, (CAGA)12-MLP-Luc reporter construct (pCAGA12-luc) and pPAI-1-Luc are generous gifts of Dr. ten Dijke P (26). FLAG-SMAD3 N and LC constructs were generous gifts from Dr. Hyunjung Ha (49). pRetroSuper-GFP SMAD3 shRNA plasmids (50) (The Addgene plasmid 15723). (See Supplementary Methods)
Targeted Disruption of Dear1 in the Mouse
DEAR1 null mice were generated by deletion of the 5' end of exon1, containing the first ATG, via replacement with a neomycin selection cassette. Following germline transmission, mice were intercrossed to maintain an identical background across genotypes. All mice used in this study were littermates. (See Supplementary Methods).
3D culture and time-lapse
Three dimensional (3D) culture assays for acini formation were performed as described by Debnath et al (46). For time-lapse, cells were plated on the top of matrigel (Invitrogen). After 6 hours incubation, cells were monitored under the invert phase microscope. Images were captured at 30 min intervals for 72 hrs in 5–7 different fields of each sample simultaneously. Recorded images were stacked as movies and average migration distances determined for at least 30 cells per field. Cell migration distance was counted and measured using the MetaMorph Program.
GST pull-down assay
GST and GST-DEAR1 fusion protein were expressed in BL21 cells and purified according to the manufacturer’s instructions (GE Health Sciences). For the GST-pull down assay, HEK293T cells were transfected with Flag-SMAD3 for 24h and then lysed in RIPA lysis buffer. The lysates in GST-binding buffer (20mM Tris-HCl pH 8.0, 0.1M NaCl, 0.2mM EDTA) were precleared by incubating with control GST and glutathione-sepharose beads for 2h at 4°C. Equal amounts of precleared lysates were then incubated with either GST-DEAR1 or control GST for 2h at 4°C. The beads were washed five times with GST-binding buffer and then eluted in 30 ul of 2xSDS-sample buffer and then detected by immunoblotting.
In vivo ubiquitination assay
For in vivo ubiquitination assay HEK293T cells were transfected with the indicated plasmids. At 20h after transfection, cells were treated with MG132 20uM for 4h and harvested. Cells were lysed in SDS-lysis buffer by boiling for 10 min and then diluted 10-fold in 0.5% NP-40 buffer and used for further analysis.
Immunofluorescence staining
Exponentially growing, cells on glass were fixed with 4% formaldehyde and permeabilized with 0.3% Triton X-100 in PBS before incubation with blocking buffer and antibodies. Stained cells were mounted with cover slips by anti-fade fluorescent mounting medium (ProLong Gold) and observed and recorded by deconvolutional microscopy. Co-localization experiments using confocal microsopy and the Imaris imaging system were carried out as described (28).
Wound assays
Cells from DEAR1 knockdown (KD) and its control clones (2×105/well) were seeded in 6-well plates. Cells were incubated for 24 hours until confluent. The wounds were made by scraping with 200ul plastic tip across the cell monolayer. The wounded cells were treated or untreated with 2 ng/ml TGFβ. Phase contrast images were recorded after 1–2 days culture.
Anoikis assays
Freshly trypsinized cells (4 × 105 /dish) were seeded into ultra-low attachment (ULA) 60mm cell culture dishes (Costar). ULA dishes with a covalently bound hydrogel layer that effectively inhibits cellular attachment. After 4 days culture, suspension cells were collected and plated to a regular 100 mm dish for colony formation. Also, the suspension cells were collected and lysed by 1x SDS sample buffer for Western blotting. Plates were stained by crystal violet and scanned with an Epson Perfection 4990 photo scanner 4 days later.
Database Analysis, RNA Extraction and Q-RT-PCR assay
Statistical analysis
All data are expressed as means with SDs. The statistical significance of differences between two groups was analyzed by two tail Student’s t tests. Differences with P values of <0.05 were considered statistically significant.
Supplementary Material
SIGNIFICANCE.
Cumulative results provide compelling evidence that DEAR1 is a critical tumor suppressor involved in multiple human cancers and provide a novel paradigm for regulation of TGFβ-induced EMT through DEAR1’s regulation of SMAD3 protein levels. DEAR1 loss of function has important therapeutic implications for targeted therapies aimed at the TGFβ/SMAD3 pathway.
Acknowledgements
The research described herein was supported by the NCI Early Detection Research Network grant (5U01CA111302-08) to AMK, SS, and MLF, the Department of Defense grant (BC111524) to NC and AMK., as well as the Metastasis Research Center, U.T. M. D. Anderson Cancer Center to AMK, the Rosalie B. Hite Fellowship, the American Legion Auxiliary scholarship and the PEO Scholars award to JR. We would like to thank Dr. John Parant for assistance with targeting construct design, and GL for initial support for electroporation of construct (NCI grant to GL). We would also like to thank Dr. Richard Behringer for assistance with the IHC analysis of tumors from our mouse model as well as Dr. Mien-Chie Hung (MD Anderson Cancer Center) for critical reading of the manuscript. We would like to thank Hank Adams for microscopy technical support. We would like to acknowledge the Genetically Engineered Mouse Facility for help with the electroporation of constructs as well as the DNA extraction core and the DNA Analysis Core for genotyping of knockout mouse and Q-RTPCR support, and Histology core for IHC staining, the TCGA project groups involved in the compilation of the TCGA data associated with breast carcinoma, colon and rectal carcinoma, squamous cell lung carcinoma, endometrial carcinoma, clear cell renal carcinoma, and glioblastoma.
Footnotes
The authors disclose no potential conflicts of interest.
Reference List
- 1.Hilgers W, Tang DJ, Sugar AY, Shekher MC, Hruban RH, Kern SE. High-resolution deletion mapping of chromosome arm 1p in pancreatic cancer identifies a major consensus region at 1p35. Genes Chromosomes Cancer. 1999;24:351–355. [PubMed] [Google Scholar]
- 2.Leister I, Weith A, Bruderlein S, Cziepluch C, Kangwanpong D, Schlag P, et al. Human Colorectal Cancer: High Frequency of Deletions at Chromosome 1p35. Cancer Res. 1990;50:7232–7235. [PubMed] [Google Scholar]
- 3.Ragnarsson G, Eiriksdottir G, Johannsdottir JT, Jonasson JG, Egilsson V, Ingvarsson S. Loss of heterozygosity at chromosome 1p in different solid human tumours: association with survival. Br J Cancer. 1999;79:1468–1474. doi: 10.1038/sj.bjc.6690234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borg A, Zhang QX, Olsson H, Wenngren E. Chromosome 1 alterations in breast cancer: allelic loss on 1p and 1q is related to lymphogenic metastases and poor prognosis. Genes Chromosomes Cancer. 1992;5:311–320. doi: 10.1002/gcc.2870050406. [DOI] [PubMed] [Google Scholar]
- 5.Ogunbiyi OA, Goodfellow PJ, Gagliardi G, Swanson PE, Birnbaum EH, Fleshman JW, et al. Prognostic value of chromosome 1p allelic loss in colon cancer. Gastroenterology. 1997;113:761–766. doi: 10.1016/s0016-5085(97)70169-0. [DOI] [PubMed] [Google Scholar]
- 6.Lott ST, Chen N, Chandler DS, Yang Q, Wang L, Rodriguez M, et al. DEAR1 Is a Dominant Regulator of Acinar Morphogenesis and an Independent Predictor of Local Recurrence-Free Survival in Early-Onset Breast Cancer. PLoS Med. 2009;6:e1000068. doi: 10.1371/journal.pmed.1000068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, et al. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muthuswamy SK. A New Tumor Suppressor That Regulates Tissue Architecture. PLoS Med. 2009;6:e1000073. doi: 10.1371/journal.pmed.1000073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wodarz A, Nathke I. Cell polarity in development and cancer. Nat Cell Biol. 2007;9:1016–1024. doi: 10.1038/ncb433. [DOI] [PubMed] [Google Scholar]
- 11.Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 12.Meulmeester E, ten Dijke P. The dynamic roles of TGF-β in cancer. J Pathol. 2011;223:206–219. doi: 10.1002/path.2785. [DOI] [PubMed] [Google Scholar]
- 13.Berger AH, Knudson AG, Pandolfi PP. A continuum model for tumour suppression. Nature. 2011;476:163–169. doi: 10.1038/nature10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J, Haider S, Baran J, Cros A, Guberman JM, Hsu J, et al. BioMart: a data federation framework for large collaborative projects. Database. 2011;2011 doi: 10.1093/database/bar038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rudin CM, Durinck S, Stawiski EW, Poirier JT, Modrusan Z, Shames DS, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet. 2012;44:1111–1116. doi: 10.1038/ng.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forer L, Schonherr S, Weissensteiner H, Haider F, Kluckner T, Gieger C, et al. CONAN: copy number variation analysis software for genome-wide association studies. BMC Bioinformatics. 2010;11:318. doi: 10.1186/1471-2105-11-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Termen S, Tan E-J, Heldin CH, Moustakas A. p53 regulates epithelial mesenchymal transition induced by transforming growth factor β. J Cell Physiol. 2013;228:801–813. doi: 10.1002/jcp.24229. [DOI] [PubMed] [Google Scholar]
- 19.Arima Y, Inoue Y, Shibata T, Hayashi H, Nagano O, Saya H, et al. Rb Depletion Results in Deregulation of E-Cadherin and Induction of Cellular Phenotypic Changes that Are Characteristic of the Epithelial-to-Mesenchymal Transition. Cancer Res. 2008;68:5104–5112. doi: 10.1158/0008-5472.CAN-07-5680. [DOI] [PubMed] [Google Scholar]
- 20.Lee JM, Dedhar S, Kalluri R, Thompson EW. The epithelial-mesenchymal transition: new insights in signaling, development, and disease. J Cell Biol. 2006;172:973–9781. doi: 10.1083/jcb.200601018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamada KM, Cukierman E. Modeling tissue morphogenesis and cancer in 3D. Cell. 2007;130:601–610. doi: 10.1016/j.cell.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 22.Yeh YC, Wei WC, Wang YK, Lin SC, Sung JM, Tang MJ. Transforming Growth Factor-{beta}1 Induces Smad3-Dependent {beta}1 Integrin Gene Expression in Epithelial-to-Mesenchymal Transition during Chronic Tubulointerstitial Fibrosis. Am J Pathol. 2010;177:1743–1754. doi: 10.2353/ajpath.2010.091183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moustakas A, Heldin CH. Dynamic control of TGF-beta signaling and its links to the cytoskeleton. FEBS Lett. 2008;582:2051–2065. doi: 10.1016/j.febslet.2008.03.027. [DOI] [PubMed] [Google Scholar]
- 24.Liotta LA, Kohn E. Anoikis: Cancer and the homeless cell. Nature. 2004;430:973–974. doi: 10.1038/430973a. [DOI] [PubMed] [Google Scholar]
- 25.Schmelzle T, Mailleux AA, Overholtzer M, Carroll JS, Solimini NL, Lightcap ES, et al. Functional role and oncogene-regulated expression of the BH3-only factor Bmf in mammary epithelial anoikis and morphogenesis. Proceedings of the National Academy of Sciences. 2007;104:3787–3792. doi: 10.1073/pnas.0700115104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S, Gauthier JM. Direct binding of Smad3 and Smad4 to critical TGF[beta]-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 1998;17:3091–3100. doi: 10.1093/emboj/17.11.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lonn P, Moren A, Raja E, Dahl M, Moustakas A. Regulating the stability of TGFbeta receptors and Smads. Cell Res. 2009;19:21–35. doi: 10.1038/cr.2008.308. [DOI] [PubMed] [Google Scholar]
- 28.Marvizon JCG, Perez OA, Song B, Chen W, Bunnett NW, Grady EF, et al. Calcitonin receptor-like receptor and receptor activity modifying protein 1 in the rat dorsal horn: Localization in glutamatergic presynaptic terminals containing opioids and adrenergic [alpha]2C receptors. Neuroscience. 2007;148:250–265. doi: 10.1016/j.neuroscience.2007.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jinnin M, Ihn H, Tamaki K. Characterization of SIS3, a Novel Specific Inhibitor of Smad3, and Its Effect on Transforming Growth Factor-β1-Induced Extracellular Matrix Expression. Mol Pharmacol. 2006;69:597–607. doi: 10.1124/mol.105.017483. [DOI] [PubMed] [Google Scholar]
- 30.Sanchez-Tillo E, Liu Y, de BO, Siles L, Fanlo L, Cuatrecasas M, et al. EMT-activating transcription factors in cancer: beyond EMT and tumor invasiveness. Cell Mol Life Sci. 2012;69:3429–3456. doi: 10.1007/s00018-012-1122-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee K, Nelson CM. New insights into the regulation of epithelial-mesenchymal transition and tissue fibrosis. Int Rev Cell Mol Biol. 2012;294:171–221. doi: 10.1016/B978-0-12-394305-7.00004-5. [DOI] [PubMed] [Google Scholar]
- 32.Sato M, Muragaki Y, Saika S, Roberts AB, Ooshima A. Targeted disruption of TGF-beta1/Smad3 signaling protects against renal tubulointerstitial fibrosis induced by unilateral ureteral obstruction. J Clin Invest. 2003;112:1486–1494. doi: 10.1172/JCI19270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shih JY, Yang PC. The EMT regulator slug and lung carcinogenesis. Carcinogenesis. 2011;32:1299–1304. doi: 10.1093/carcin/bgr110. [DOI] [PubMed] [Google Scholar]
- 34.Su X, Chakravarti D, Cho MS, Liu L, Gi YJ, Lin YL, et al. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature. 2010;467:986–990. doi: 10.1038/nature09459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Venkatachalam S, Shi YP, Jones SN, Vogel H, Bradley A, Pinkel D, et al. Retention of wild-type p53 in tumors from p53 heterozygous mice: reduction of p53 dosage can promote cancer formation. EMBO J. 1998;17:4657–4667. doi: 10.1093/emboj/17.16.4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santarosa M, Ashworth A. Haploinsufficiency for tumour suppressor genes: when you don't need to go all the way. Biochimica et Biophysica Acta (BBA) - Reviews on Cancer. 2004;1654:105–122. doi: 10.1016/j.bbcan.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 37.Kaghad M, Bonnet H, Yang A, Creancier L, Biscan JC, Valent A, et al. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997;90:809–819. doi: 10.1016/s0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- 38.Bagchi A, Papazoglu C, Wu Y, Capurso D, Brodt M, Francis D, et al. CHD5 is a tumor suppressor at human 1p36. Cell. 2007;128:459–475. doi: 10.1016/j.cell.2006.11.052. [DOI] [PubMed] [Google Scholar]
- 39.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Remy I, Montmarquette A, Michnick SW. PKB/Akt modulates TGF-[beta] signalling through a direct interaction with Smad3. Nat Cell Biol. 2004;6:358–365. doi: 10.1038/ncb1113. [DOI] [PubMed] [Google Scholar]
- 41.Conery AR, Cao Y, Thompson EA, Townsend C, Ko TC, Luo K. Akt interacts directly with Smad3 to regulate the sensitivity to TGF-[beta]-induced apoptosis. Nat Cell Biol. 2004;6:366–372. doi: 10.1038/ncb1117. [DOI] [PubMed] [Google Scholar]
- 42.Guo X, Ramirez A, Waddell DS, Li Z, Liu X, Wang XF. Axin and GSK3-β control Smad3 protein stability and modulate TGF-β signaling. Genes Dev. 2008;22:106–120. doi: 10.1101/gad.1590908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bachman KE, Park BH. Duel nature of TGF-[beta] signaling: tumor suppressor vs. tumor promoter. Current Opinion in Oncology. 2005:17. doi: 10.1097/01.cco.0000143682.45316.ae. [DOI] [PubMed] [Google Scholar]
- 44.Millet C, Zhang YE. Roles of Smad3 in TGF-beta signaling during carcinogenesis. Crit Rev Eukaryot Gene Expr. 2007;17:281–293. doi: 10.1615/critreveukargeneexpr.v17.i4.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagaraj NS, Datta PK. Targeting the transforming growth factor-beta signaling pathway in human cancer. Expert Opinion on Investigational Drugs. 2010;19:77–91. doi: 10.1517/13543780903382609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 47.Lee MT, Coburn GA, McClure MO, Cullen BR. Inhibition of Human Immunodeficiency Virus Type 1 Replication in Primary Macrophages by Using Tat- or CCR5-Specific Small Interfering RNAs Expressed from a Lentivirus Vector. J Virol. 2003;77:11964–11972. doi: 10.1128/JVI.77.22.11964-11972.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilkinson DS, Tsai WW, Schumacher MA, Barton MC. Chromatin-bound p53 anchors activated Smads and the mSin3A corepressor to confer transforming-growth-factor-beta-mediated transcription repression. Mol Cell Biol. 2008;28:1988–1998. doi: 10.1128/MCB.01442-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seong HA, Jung H, Ha H. Murine protein serine/threonine kinase 38 stimulates TGF-beta signaling in a kinase-dependent manner via direct phosphorylation of Smad proteins. J Biol Chem. 2010;285:30959–30970. doi: 10.1074/jbc.M110.138370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He W, Dorn DC, Erdjument-Bromage H, Tempst P, Moore MAS, Massague J. Hematopoiesis Controlled by Distinct TIF1β and Smad4 Branches of the TGFβ Pathway. Cell. 2006;125:929–941. doi: 10.1016/j.cell.2006.03.045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.