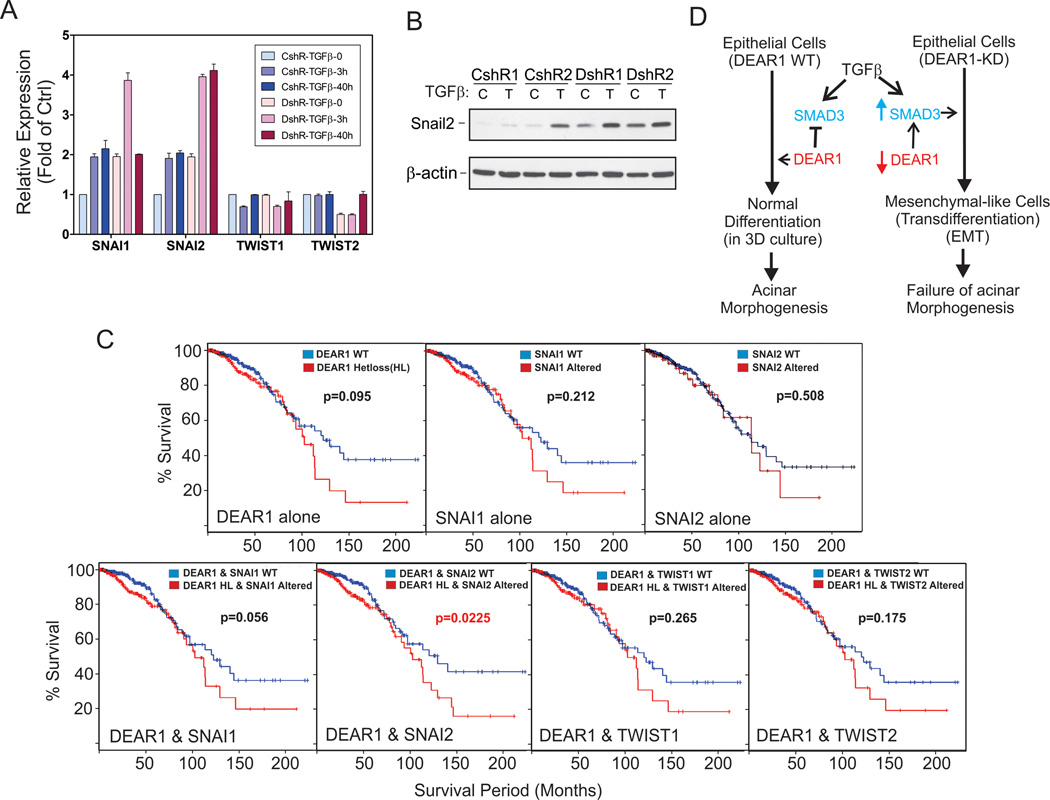
Figure 7. (A) Quantitative RT-PCR analysis of SNAI1/2 in DEAR1-shRNA knockdown clones.
76NE6 DEAR1-KD cells were treated with or without TGFβ (4ng/ml) for 3h and 40h, then, were collected in TRIzol. RNA were extracted following TRIzol instruction and purified with Absolutely RNA microprep kit (Stratagene). Q-PCR was performed using TaqMan (Applied Biosystems). (B) Western analysis of Snail2 (Slug). 76N-E6 DEAR1-KD (DshR1 and DshR2) and control clones (CshR1 and CshR2) were treated with or without TGFβ (4 ng/ml) for 1 day. (C) The effect of DEAR1 heterozygous loss and SNAI1/2 gene upregulation on survival of invasive breast cancer patients. Survival curves were generated by cBio, using Kaplan-Meier analysis through querying complete tumor sets in the BRCA cohort for DEAR1 heterozgyous loss, SNAI1, SNAI2, TWIST1 and TWIST2. Alteration of SNAI1/2 and TWIST1/2 includes amplification, upregulation of mRNA/protein expression (if applicable) greater than two standard deviations from the mean. (D) A novel model for the regulation of TGFβ-induced EMT by DEAR1