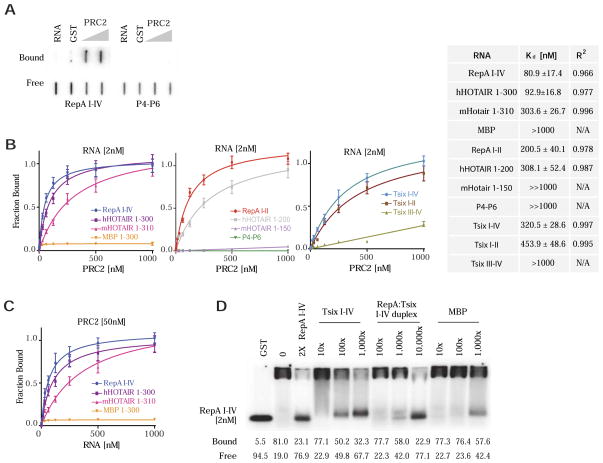
Figure 2. A range of PRC2-RNA binding affinities.
(A) Example of double-filter binding assay used to obtain binding isotherms and Kd values. Top (bound), nitrocellulose membrane. Bottom (free), Hybond-N+ membrane. (B) Binding isotherms of PRC2 to 2 nM RNA of species indicated. Equilibrium dissociation constants (Kd) and R2 values shown in table. N/A, not applicable because curves were too flat. “≫1000 nM” denotes a Kd in excess of that which could be measured under our assay conditions. (C) Reciprocal binding experiments in which 1–1000nM RNA is titrated against constant 50nM PRC2, with Kd and R2 values shown. A labeled corresponding RNA (<1%) was used as tracer. (D) EMSA using 2nM RepA I-IV probe in the presence of cold competitor RNA species (RepA I-IV, Tsix I-IV, RepA I-IV: Tsix I-IV pre-annealed duplex, MBP) at indicated concentrations.