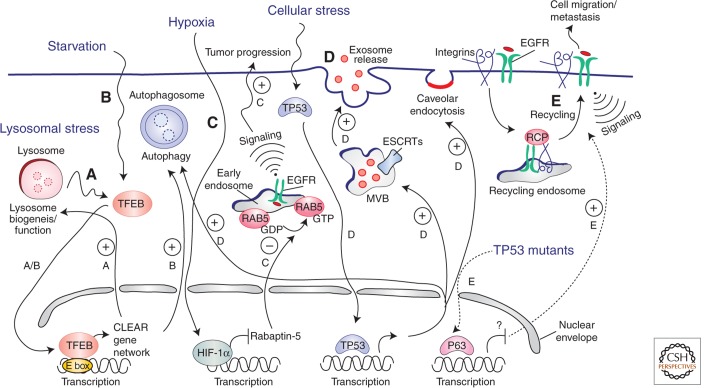
Figure 1.
Transcriptional programs controlling endocytosis. Lysosomal stress (A) and/or starvation (B) cause nuclear translocation of the TFEB transcription factor, which binds to the E-box sequence on promoters and induces transcription of a cluster of genes (the “CLEAR network”) involved in lysosomal biogenesis/function and autophagy genes, promoting cellular clearance of nondegraded molecules. (C) Activation of HIF1 during hypoxia causes the inhibition of the RABAPTIN-5 gene transcription, inhibition of Rab5 GTP-loading and, consequently, endosomal retention of the EGFR, eventually leading to sustained EGFR signaling from the endosomal station and tumor progression. (D) On different types of cellular stresses, p53 translocates into the nucleus and activates the transcription of genes that play roles at different stations of the endocytic pathway: autophagy genes (e.g., lysosome/autophagosome components), genes involved in exosome release from MVBs (e.g., ESCRT components), and the CAV-1 gene, which stimulates caveolar endocytosis. (E) TP53 mutations found in human cancers exert part of their oncogenic potential through the P63-dependent (transcriptional-dependent) stimulation of RCP-mediated recycling of integrin-EGFR complexes, leading to induced migration and metastasis. The molecular mechanism for this remains unclear.