Figure 1.
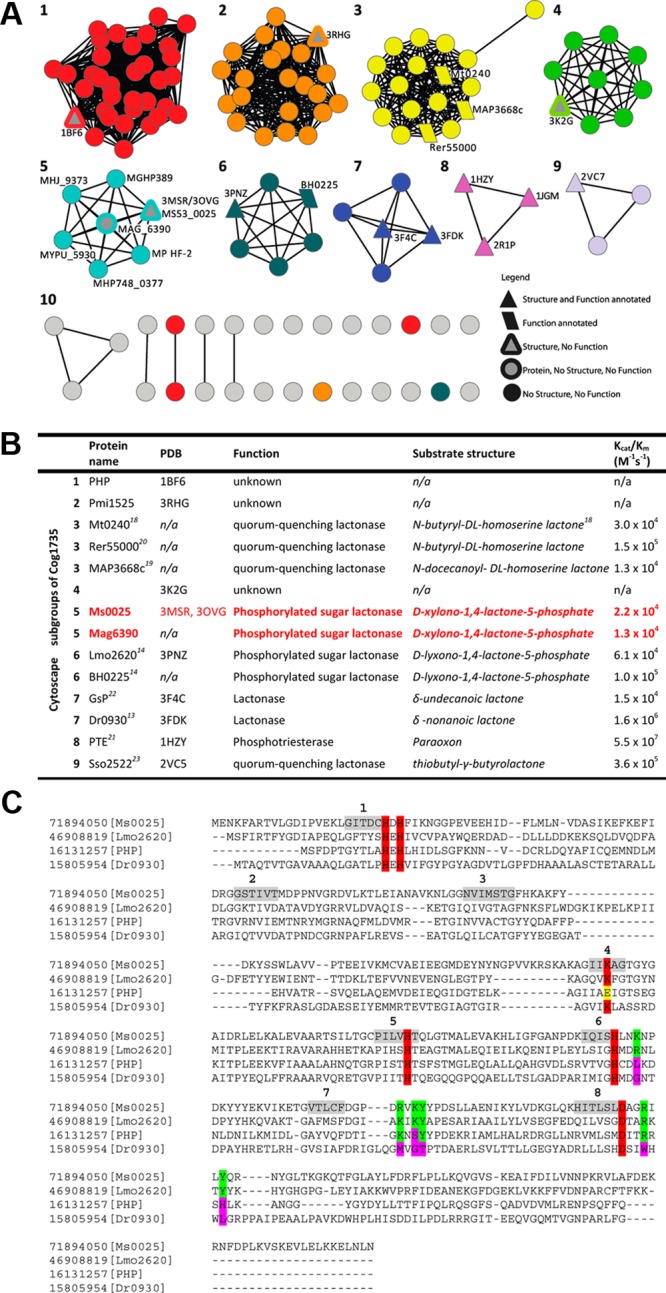
Sequence similarity network organization showing one of the 24 clusters representing the amidohydrolase superfamily sequences annotated as belonging to Cog1735. (A) Each node in the diagram represents a sequence, and each edge represents the pairwise connection between two sequences at a BLAST E value of better than 10–80. All functionally annotated proteins (trapezoids) as well as structurally characterized proteins (triangles) have been mapped onto the nodes. Ms0025 (cyan and gray triangle) and Mag6390 (cyan and gray circle) are found in subgroup 6. (B) List summarizing each of the known proteins, structures, substrate profiles, and kinetic parameters for each node. Colored red are Ms0025 and Mag6390, which are the phosphorylated sugar lactonases that are able to hydrolyze d-xylono-1,4-lactone-5-phosphate. (C) Structure-based sequence alignment of Ms0025 (gi|71894050, subgroup 6), Lmo2620 (gi|46908819, subgroup 5), phosphotriesterase homology protein (gi|16131257, subgroup 1), and Dr0930 (gi|15805954, group 7). The amino acids required for metal binding are highlighted in red. The residues implicated in phosphate binding are highlighted in green and the corresponding residues in the three other Cog1735 proteins in pink. Regions highlighted in gray represent the strands of the barrel fold.
