Figure 5.
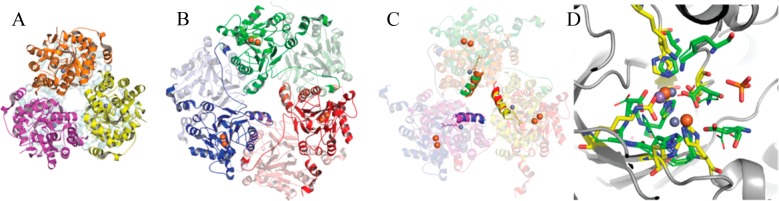
Comparison of lactonase structures of Ms0025 (PDB entry 3OVG) and UlaG (PDB entry 3BV6/2WYL). (A) Ribbon representation of the hexamer of Ms0025 that is seen in the crystal structure as a dimer (light blue) of trimers (yellow, pink, and orange). The metal ions at each active site are shown as gray spheres. The molecules are oriented with the 3-fold axis perpendicular to the plane of the page. (B) UlaG hexamer viewed down the molecular 3-fold axis, which is a trimer of dimer (red, blue, and green and dimer chain in light colors). (C) Superposition of Ms0025 and UlaG based on the superposition of the helix around the central solvent pore in UlaG (green → orange, red → yellow, blue → pink). The two zinc atoms for each protein are depicted as spheres and show a different position orientation of the active site. (D) Superposition of the holoenzyme active sites of Ms0025 and UlaG. Active site residues are represented as yellow sticks for Ms0025 and green sticks for UlaG, with Zn2+ shown as gray spheres and Mn2+ and Fe3+ shown as purple and orange spheres, respectively. In addition to metal-coordinating residues, the location of the side chain of Lys29 (Ms0025) and Lys260 (UlaG) is shown, which appears to be in a favorable position for substrate interaction, albeit coming from a different direction.
