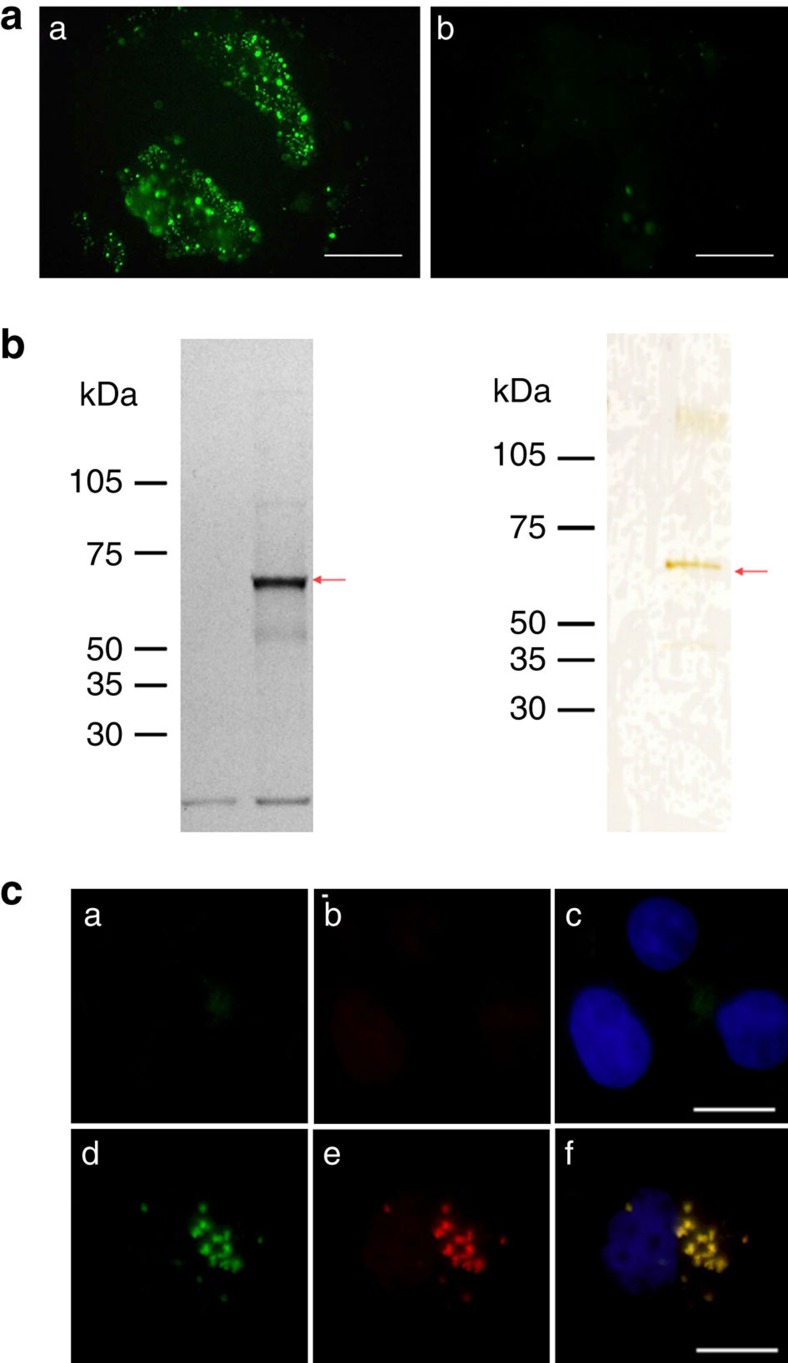
Figure 2. Identification of z13 peptide receptor.
(a) Fluorescence micrographs of Ishikawa cells (left) and control A431 cells (right) overlayed with a synthetic z13 peptide tagged with fluorescein isothiocyanate (FITC) and left at 37 °C for 15 min. Scale bar, 50 μm. (b) Visualization and isolation of the z13 peptide receptor. Left: cell surface proteins expressed on Ishikawa cells were biotinylated. Cell lysates were bound to z13 peptide-conjugated agarose beads, and bound proteins were eluted with irrelevant peptide (lane 1) or z13 peptide (lane 2). Biotinylated proteins in each eluate were detected by peroxidase-conjugated avidin and a luminescent peroxidase substrate. Right: silver staining of peptide-affinity-purified z13 receptor from endometriosis. Endometriosis tissues isolated from patients were homogenized, and microsome membrane fraction was prepared. Proteins solubilized with detergent were applied to a z13 peptide-conjugated agarose column, and bound proteins were eluted with irrelevant peptide (lane 1) or z13 peptide (lane 2). Proteins in each eluate in SDS–PAGE were detected by silver staining. (c) Fluorescence micrographs of HeLa cells transfected with control empty vector (upper row) or with an expression vector encoding CNGB3–MYC (lower row). Binding of FITC-z13 peptide (green) to HeLa cells transfected with mammalian expression vectors (a,d), immunostained with anti-MYC followed by Alexa 549 (red)-conjugated anti-mouse IgG antibody (b,e) and merged images including 4',6-diamidino-2-phenylindole (blue) to indicate nuclear staining (c,f). Scale bar, 20 μm.