Figure 1. ChaC1 efficiently and selectively degrades reduced glutathione.
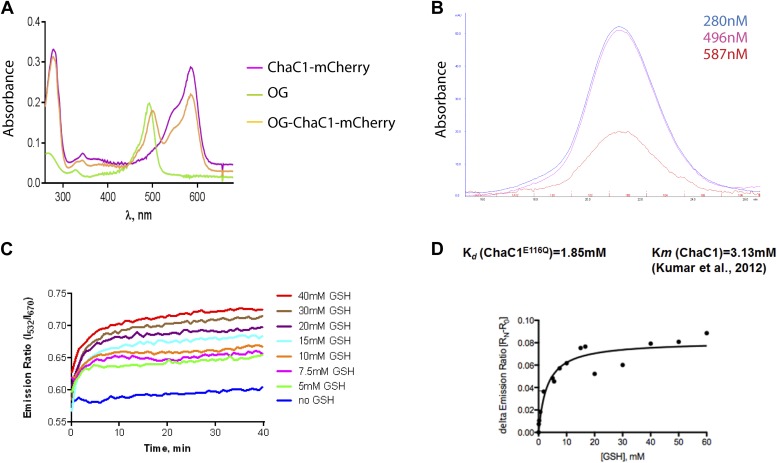
(A) A bar-graph representation of residual glutathione levels following incubation of 10 mM glutathione with the indicated concentrations of bacterially expressed mouse ChaC1. Varying concentrations of enzymes were assayed at a single time point (left panel) and varying initial concentrations of glutathione were assayed at different time points (right panel). (B) Comparison of the ability of ChaC1 to eliminate reduced (GSH) and oxidized glutathione (GSSG). Note that ChaC1 effectively eliminated reduced glutathione, but had no effect on oxidized glutathione. (C) Cartoon of the fluorescent resonance energy transfer (FRET) probe, OG-ChaC1-Cherry, used to detect substrate binding to ChaC1. Shown is a model of murine ChaC1 (UniProt Q8R3J5) residues 31–204, created by Phyre2 (Kelley and Sternberg, 2009) based on the crystal structure of γ-glutamyl cyclotransferase (PDB 2RBH). The side chain of Cys 92, which has been modified with the Oregon Green (OG) donor, is highlighted, as is the C-terminus of the protein, site of the fused mCherry fluorescent acceptor. (D) Time-resolved FRET signal (expressed as the ratio of the emission signal at 532 nm and 670 nm upon excitation at 480 nm) of the OG-ChaC1-Cherry probe [2.5 µM] following exposure to 10 mM reduced (GSH) or oxidized glutathione(GSSG). Where indicated, the sample was injected with dithiotreitol (DTT) to reduce the GSSG and convert it to a substrate for ChaC1. The biphasic change in FRET signal upon exposure to GSH is consistent with binding followed by breakdown of GSH by the probe, which retains its enzymatic activity. (E) Comparison of glutathione elimination by purified bacterially expressed wild-type and E116Q mutant ChaC1 in vitro.