Abstract
A wide range of antiviral drugs is currently available; however, drug-resistant viruses have begun to emerge and represent a potential public health risk. Here, we explored the use of compounds that inhibit or interfere with the action of essential host factors to prevent virus replication. In particular, we focused on the cyclin-dependent kinase 9 (CDK9) inhibitor, FIT-039, which suppressed replication of a broad spectrum of DNA viruses through inhibition of mRNA transcription. Specifically, FIT-039 inhibited replication of herpes simplex virus 1 (HSV-1), HSV-2, human adenovirus, and human cytomegalovirus in cultured cells, and topical application of FIT-039 ointment suppressed skin legion formation in a murine HSV-1 infection model. FIT-039 did not affect cell cycle progression or cellular proliferation in host cells. Compared with the general CDK inhibitor flavopiridol, transcriptome analyses of FIT-039–treated cells revealed that FIT-039 specifically inhibited CDK9. Given at concentrations above the inhibitory concentration, FIT-039 did not have a cytotoxic effect on mammalian cells. Importantly, administration of FIT-039 ameliorated the severity of skin lesion formation in mice infected with an acyclovir-resistant HSV-1, without noticeable adverse effects. Together, these data indicate that FIT-039 has potential as an antiviral agent for clinical therapeutics.
Introduction
The number of available antiviral drugs has increased continually since the development of azidothymidine and acyclovir (ACV) in the 1970s (1, 2). However, this has resulted in the emergence of drug-resistant viruses, including HIV, hepatitis B, and the herpes simplex virus (HSV), which cause drug failures (3, 4). Although the drug resistance of HSV to ACV and related drugs, such as famciclovir or valacyclovir, rarely occurs in immunocompetent individuals, it has been more commonly reported in immunocompromised patients, including patients with HIV and transplant recipients (5). A focus on host cellular proteins (6–8) is necessary to overcome drug resistance to antiviral agents, because viruses often change their genomic sequences and acquire drug resistance (4). We have developed chemical inhibitors that target host kinases as antiviral agents, which prevent the replication of several viruses, including the hepatitis C virus, dengue virus, and Sindbis virus (9–12). To develop novel drugs, we have been recently focusing on cyclin-dependent protein kinases (CDKs), which are known to be used for virus replication (13–16).
CDKs comprise a family of serine/threonine kinases that play essential roles in the regulation of multiple cellular processes and can be divided into two major groups: CDK1, CDK2, CDK3, CDK4, and CDK6 are directly involved in cell cycle regulation, while CDK5, CDK7, CDK8, CDK9, CDK11, CDK12, and CDK13 have been implicated in the control of gene transcription (17, 18). The virus-specific regulation of CDK activity has been reported, as described below. CDK1 was shown to activate the expression of HSV-1 late genes (19), while CDK2 promoted the replication of adenovirus, papillomavirus, and herpes virus DNAs (7). CDK7 and CDK9 were shown to activate viral RNA transcription in HIV and HSV (7, 14, 15, 20–24). Feichtinger et al. recently described the interaction between CDK9/cyclin T1 and viral mRNA export factor pUL69 in human cytomegalovirus (HCMV), which belongs to the β subgroup of Herpesviridae (20). In mammalian cells, CDK9/cyclin T1 (or T2) is part of the subunit of the positive transcription elongation factor b (P-TEFb) complex and reportedly initiated the transcriptional elongation of genes by phosphorylating the carboxyterminal domain (CTD) of RNA polymerase II at Ser2 (25). These findings indicate that CDKs are also used in viral DNA replication and transcription.
Flavopiridol is a pan-specific CDK inhibitor that has been shown to inhibit CDK1, CDK2, CDK4, CDK5, CDK6, CDK7, and CDK9 and suppress the replication of HSV, HCMV, human adenovirus (HAdV), and HIV (15). Roscovitine, an oligo-specific CDK inhibitor that inhibits CDK1, CDK2, CDK5, CDK7, CDK9, and CDK12, suppresses the transcription of immediate-early genes (IEGs) of HCMV and prevents viral replication in vitro (14, 26–28). However, these broad-spectrum CDK inhibitors often negatively affect cell cycle progression and elicit adverse effects, which lead to difficulties in the development of antiviral drugs (29).
In this study, we developed the CDK9 inhibitor, FIT-039, which suppressed the replication of HSV and other DNA viruses, including HCMV and HAdV. It also suppressed the replication of ACV-resistant HSV-1, not only in vitro, but also in vivo. Our ongoing preclinical studies have shown no significant adverse effects. These findings suggest that FIT-039 is a promising compound for the therapeutic treatment of drug-resistant HSVs and other DNA viruses.
Results
Requirement of CDK9 for HSV-1 replication but not for host cell cycle progression.
To establish whether CDK9 was required for HSV-1 replication, we investigated the replication level of HSV-1 in HeLa cells transfected with CDK9 siRNA or scramble dsRNA as a negative control (Supplemental Figure 1A; supplemental material available online with this article; doi:10.1172/JCI73805DS1). The cells were infected with HSV-1 at a MOI of 0.1 PFU per cell, and the copy number of HSV-1 genomic DNA was measured using real-time PCR. Knockdown of CDK9 significantly suppressed the replication of HSV-1 genomic DNA (Supplemental Figure 1B), which demonstrated the necessity for CDK9 in the replication of HSV-1. In addition, knockdown of CDK9 had a similar effect on cell cycle progression to that of the negative control, whereas CDK4 reduced the number of S-phase cells (Supplemental Figure 1, C and D). These results support our hypothesis that CDK9 is a potential target for anti–HSV-1 drug discovery.
Identification of a CDK9-selective inhibitor.
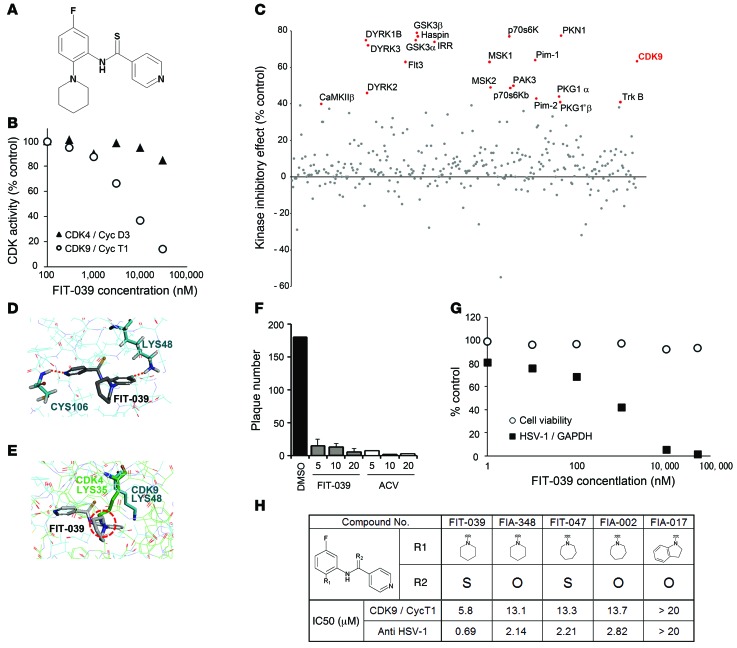
To obtain a selective inhibitor for CDK9, we performed in vitro screening assays on our original synthetic chemical library to identify small molecules that suppressed the kinase activity of CDK9 but not that of CDK4. As a result, we found a good candidate, N-[5-fluoro-2-(1-piperidinyl)phenyl]isonicotinthioamide (Figure 1A), which we refer to herein as FIT-039 (Figure 1A), which we refer to herein as FIT-039. This compound suppressed the in vitro kinase activity of CKD9/cyclin T1 with an IC50 value of 5.8 μM but did not have a marked inhibitory effect on CDK4/cyclinD3 up to 30 μM (Figure 1B). FIT-039 also did not inhibit other CDKs, including CDK2/cyclinB1, CDK2/cyclinE1, CDK5/p25, CDK6/cyclinD3, or CDK7/cyclinH/MAT (Supplemental Figure 2). We checked the effect of FIT-039 on other kinases and found that FIT-039 potently suppresses 8 kinases (GSK3β, PKN1, haspin, p70s6K, DYRK1B, GSK3α, IRR, and DYRK3) other than CDK9 on the 332-kinase panel (Figure 1C and Supplemental Table 1).
Figure 1. Identification of the CDK9-selective inhibitor FIT-039, which suppressed the replication of HSV-1.
(A) Chemical structure of FIT-039. (B) In vitro kinase assay of CDK9/cyclin T1 and CDK4/cyclinD3. An increased amount of FIT-039 inhibited CDK9/cyclin T1 but not CDK4/cyclinD3. (C) Scatter plots of the 332-kinase panel of FIT-039 (10 μM). Red dots indicate greater than 40% inhibition. (D) A structural model for FIT-039 bound to CDK9. FIT-039 formed 2 hydrogen bonds in the ATP-binding pocket, in which LYS48 and CYS106 interacted with the fluoride moiety and nitrogen atom of the pyridine of FIT-039, respectively. (E) The ATP-binding pocket of CDK4 (thin green lines) was superimposed onto that of CDK9 (thin blue lines) in the CDK9/FIT-039 complex model shown in Figure 2D. The different orientation of the side chain of LYS35 of CDK4, which corresponds to the LYS48 of CDK9, would hinder the binding of FIT-039 to CDK4. (F) FIT-039 suppressed the plaque formation caused by the HSV-1 infection in a dose-dependent manner. HSV-1 plaque reduction assay was performed in Vero cells treated with DMSO, FIT-039, and ACV. (G) FIT-039 inhibited the replication of HSV-1 genomic DNA in a dose-dependent manner without cytotoxicity. HSV-1 replication and cytotoxicity were analyzed by real-time PCR and the WST-8 assay, respectively. (H) Structure-activity relationship. The chemical structures of FIT-039 and its derivatives are shown. IC50 values of anti–HSV-1 activity were determined by real-time PCR of HSV-1 genomic DNA replication in infected HeLa cells. The IC50 values of CDK9/cyclin T1 inhibitory activity were calculated by the in vitro kinase assay. The IC50 values of anti–HSV-1 activity were correlated with those of CDK9/cyclin T1 activity (R2 = 0.92).
To examine whether FIT-039 bound to the ATP-binding pocket of CDK9, we analyzed the inhibitory action of FIT-039 on CKD9/cyclin T1 activity with a double-reciprocal Lineweaver-Burk plot, which demonstrated that FIT-039 competed with ATP for CDK9 (Supplemental Figure 3). We then performed a docking simulation to verify the binding properties of FIT-039 to the ATP-binding pocket of CDK9. LYS48 and CYS106 of CDK9 were speculated to interact with FIT-039 by forming hydrogen bonds with the fluoride group and nitrogen atom of pyridine, respectively (Figure 1D). On the other hand, LYS35 of CDK4, which corresponds to LYS48 of CDK9, appeared to serve as a steric hindrance to the binding of FIT-039 to the ATP-binding pocket of CDK4 (Figure 1E), which rationalized the selective inhibition of CDK9 by FIT-039.
FIT-039 suppressed viral replication without affecting the host cell cycle.
The anti–HSV-1 activity of FIT-039 was examined with a plaque reduction assay using Vero cells at a MOI of 100 PFUs per well. FIT-039 suppressed plaque formation in a dose-dependent manner as potently as ACV, which was used as a positive control (Figure 1F). To investigate how FIT-039 suppressed plaque formation, we first examined the effect of FIT-039 on the replication of HSV-1 genomic DNA. FIT-039 inhibited replication of the HSV-1 genome in a dose-dependent manner (EC50 and EC80 were 0.69 μM and 4.0 μM, respectively), without any adverse effect on cell viability until levels of FIT-039 reached the solubility limit (Figure 1G). Although flavopiridol also suppressed replication of the HSV-1 genome (EC50 and EC80 were 34 nM and 79 nM, respectively), cell viability was also reduced at the same dose range (data not shown). These results demonstrate that FIT-039 is a novel type of CDK9 inhibitor, which has the potential to be an anti–HSV-1 drug.
Anti–HSV-1 activity of FIT-039 was brought through CDK9 inhibition.
To assess the relationship between the inhibitory potency of FIT-039 on CKD9/cyclin T1 activity and HSV-1 replication, we synthesized some amide and thioamide analogs with different nitrogen substituents, including hexamethyleneimine and an indolinyl group, and evaluated their CDK9 inhibitory activity and anti–HSV-1 effects (Figure 1H). Substitution of the piperidinyl group at the R1 position with a larger homopiperidinyl group and of sulfur at the R2 position with oxygen decreased both the anti–HSV-1 and CDK9 inhibitory activities. On the other hand, the loss of inhibitory activity was observed when the R1 group was changed to indolinyl. The anti–HSV-1 activities of these compounds correlated with their inhibitory activities on CDK9 (R2 = 0.92), which strongly suggested that the antiviral effect of FIT-039 was triggered by the inhibition of CDK9.
FIT-039 inhibited viral transcription.
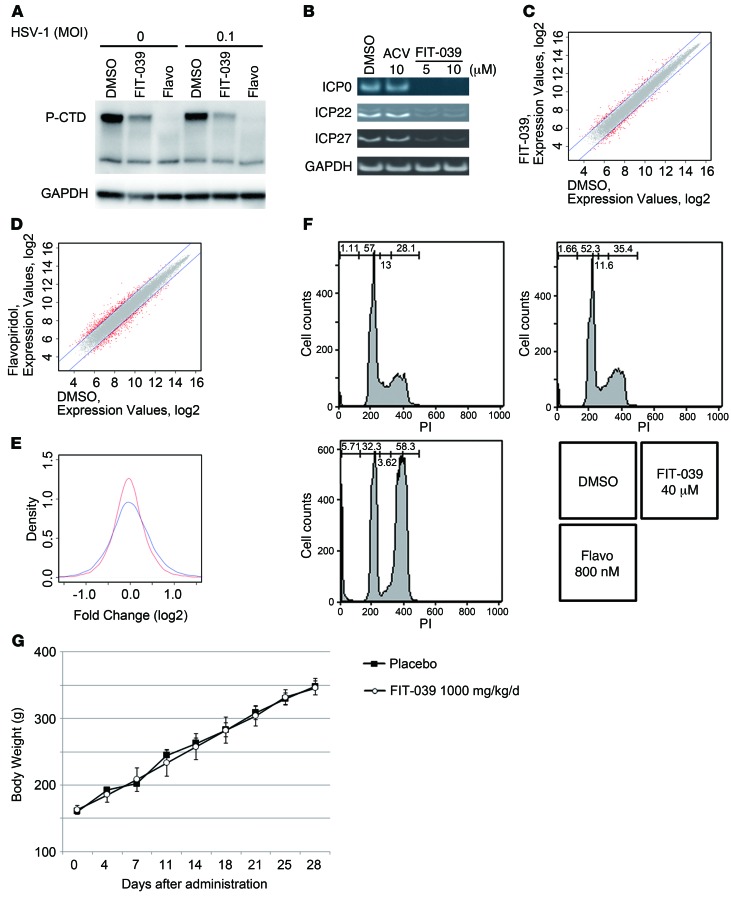
CDK9 is part of the subunit of the P-TEFb complex and promotes transcriptional elongation through phosphorylation of the CTD of RNA polymerase II (25). To elucidate the antiviral mechanism of FIT-039 on the HSV-1 replication, we investigated whether FIT-039 affects the phosphorylation of the CTD of RNA polymerase II, because several CDKs, including CDK9, phosphorylate the CTD of RNA polymerase II (18). HEK293 cells were infected with HSV-1 at a MOI of 0 or 0.1 PFUs per cell and then were treated with FIT-039 or flavopiridol for 3 hours. FIT-039 decreased phosphorylated CTD in the infected or noninfected cells to a level lower than that shown by flavopiridol (Figure 2A). Accordingly, FIT-039 reduced the expression levels of HSV-1 IEGs and early and late genes (Figure 2B and Supplemental Figure 4). IEGs are transcribed immediately after the viral infection, and their encoded proteins regulate the transcription of early and late viral genes (30). These results indicated that the antiviral mechanism may be attributed to the inhibition of the viral transcription through suppression of the CTD phosphorylation of RNA polymerase II.
Figure 2. FIT-039 inhibited the transcription of HSV-1 IEGs.
(A) HEK293 cells were infected with HSV-1 at a MOI of 0.1 PFUs per cell, and then the cells were treated with FIT-039 (30 μM) or flavopiridol (1 μM) for 3 hours. RNA polymerase II was detected by antibody against RNA polymerase II with Western blotting analysis. P-CTD, phosphorylated RNA polymerase II CTD. (B) HSV-1–attached cells were incubated for 3 hours with the indicated compounds. HSV-1 IEGs were analyzed by RT-PCR. (C and D) Overlaid scatter plots comparing mRNA expression. Two comparisons of the expression signals (C) between the FIT-039 and DMSO and (D) between the flavopiridol and DMSO treatments were analyzed. Two-fold variations in expression signals were plotted in red. (E) F tests comparing the divergences of gene expression between FIT-039 (red line) and flavopiridol (blue line). (F) HeLa cells were treated with FIT-039 and flavopiridol at a concentration 10-fold higher than that of the IC80 value of anti–HSV-1 for 24 hours. The cells were stained with propidium iodide and analyzed by flow cytometry. (G) Two-week repeated-dose oral toxicity study in rats. FIT-039 (1,000 mg/kg) or the solvent (carboxymethylcellulose) were administrated orally to male rats once a day for 14 days. Body weights and general conditions were determined for 28 days from the first administration. Flavo, flavopiridol.
Effect of FIT-039 on transcriptome of host cells.
We next evaluated the effect of FIT-039 on host cellular transcriptome. HeLa cells were incubated with FIT-039 or flavopiridol at the concentration of IC80 for 24 hours, and cellular RNAs were collected and subjected to microarray analysis. The solvent DMSO was used as a negative control. We compared divergences in gene expression between FIT-039 and flavopiridol with scatter plots (Figure 2, C and D) and F tests. We found that fold changes in FIT-039/DMSO were smaller than those in flavopiridol/DMSO. The F test showed that the difference between SDs of these 2 data sets was statistically significant (P < 10–10, Figure 2E), which implied that FIT-039 produced a less significant effect on cellular transcriptome than that of flavopiridol, whereas FIT-039 and flavopiridol similarly suppressed the viral transcription of IEGs and early and late genes of HSV-1 under these experimental conditions (Supplemental Figure 4).
To verify the difference in cytotoxicity between FIT-039 and flavopiridol, we examined the effect of these compounds on the cell cycle at a concentration 10 times higher than the IC80. FIT-039 did not affect cell cycle progression up to 40 μM, whereas flavopiridol arrested the cell cycle at the G2/M phase (Figure 2F), which demonstrated that flavopiridol caused a severe adverse effect. On the other hand, a preclinical study showed that FIT-039 did not affect body weight gain in mice administrated with an overdose of this compound (1,000 mg/kg/d) for 14 days (Figure 2G), and no changes were observed in biological markers in their blood (Supplemental Table 2).
Therapeutic activity of FIT-039 in mice.
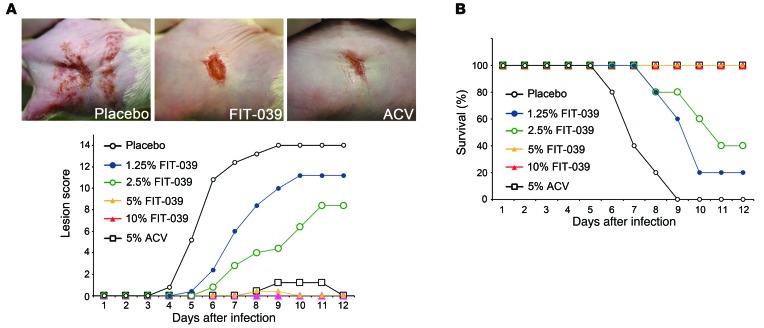
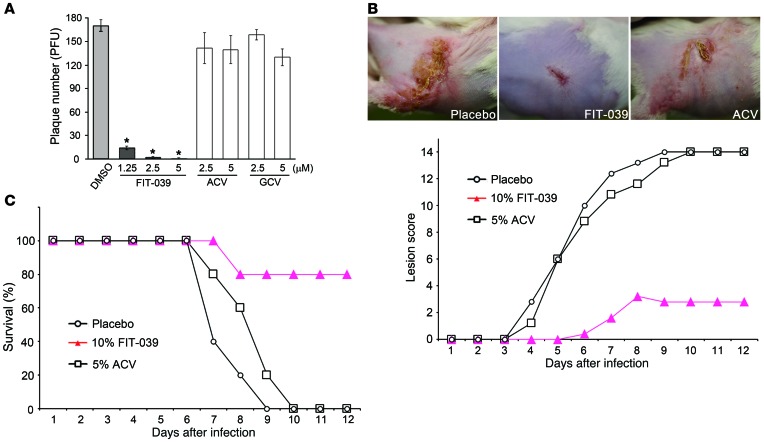
The therapeutic effect of FIT-039 was evaluated in the murine zosteriform spread model, which mimics recurrent cutaneous herpetic disease. FIT-039 ointment containing 10% lactic acid and polyethylene glycol was administrated topically to mice. ACV ointment was used as a positive control. The mid-flanks of male BALB/c mice were clipped and chemically depilated. The naked skins of mice were scratched 2 or 3 days later, and a HSV-1 suspension of 1 × 106 PFUs was then applied to the scarified area. The intradermal infection of mice at the mid-flank led to local HSV-1 replication, followed by entry of the virus into the nerves innervating the skin, and its spread to the ganglia. Following replication in the ganglia, the virus disseminated along the sensory nerves, leading to sacculation in the whole innervated dermatome after 3 to 4 days and the subsequent formation of a zosteriform lesion (31). This viral spread from sensory ganglia to the innervated skin has been used as a model for recrudescent diseases (32). The appearance of infected mice was similar to that of uninfected controls for the first 2 days after the infection. Redness in the skin began to appear on day 3 in placebo-treated mice, blisters became obvious and developed into ulcers by day 4 to 5, and all infected mice succumbed to lethal encephalitis and died on day 9.
Treatment with the FIT-039 ointment twice a day suppressed skin lesions (Figure 3A) and rescued mice from lethality (Figure 3B) in a dose-dependent manner. The healing of lesions was observed with 5% to 10% FIT-039 ointment, leading to the complete regression of zosteriform spread on day 10, which was also observed with the 5% ACV ointment. Furthermore, no skin irritation was observed with the FIT-039 treatment in mice (Figure 3A) or rabbits during a preclinical irritation test (Supplemental Figure 5). These results confirmed the CDK9 inhibitor FIT-039 as a potential candidate for a novel therapeutic drug to treat HSV-1 infections.
Figure 3. The FIT-039 ointment suppressed skin lesions caused by HSV-1 infection.
The scratched skins of mice were infected intracutaneously with HSV-1 and then treated with ointments containing the indicated concentrations of FIT-039. The ACV ointment was used as a positive control. Five mice were subjected to each experimental group. (A) The lesion score and (B) survival rate were determined. (A) Representative photographs of the infected mice 7 days after infection are shown. Each point represents the average of the results (n = 5).
Replication of a broad spectrum of DNA viruses was suppressed by FIT-039.
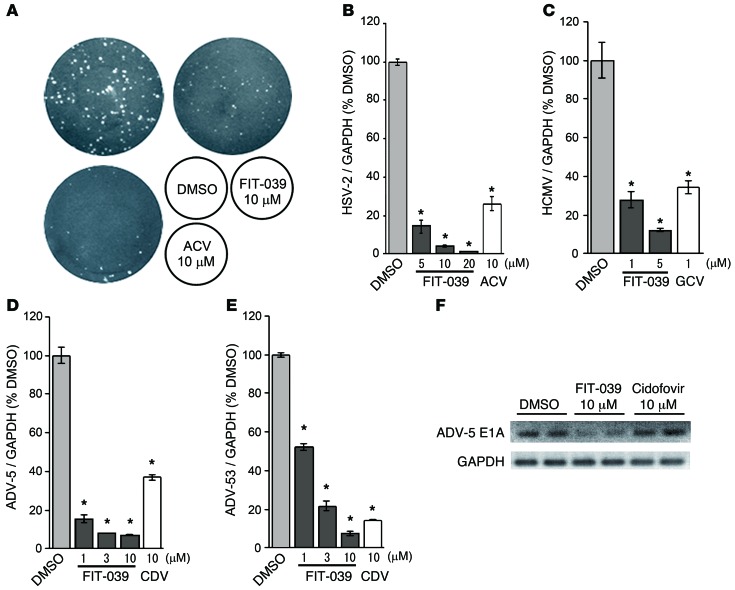
Since CDK9/cyclin T1 complex was reported previously to be required for the replication of other viruses of Herpesviridae origin, we next verified the antiviral effect of FIT-039 on HSV-2 and HCMV. FIT-039 suppressed the replication of HSV-2 in a dose-dependent manner, and its potency was similar to that of ACV (Figure 4, A and B). FIT-039 also inhibited HCMV genomic DNA replication, similar to ganciclovir (GCV) (Figure 4C). Furthermore, compared with cidofovir (CDV), FIT-039 potently inhibited the genomic DNA replication of HAdV type 5, which was obtained from a patient’s throat (Figure 4D), as well as type 53 (Figure 4E) and type 19 and type 37 (data not shown), which was obtained from a patient’s eyelid, which indicated that FIT-039 inhibited the replication of DNA viruses other than Herpesviridae. We also observed that FIT-039 reduced the expression of E1A, an early gene of HAdV, whereas CDV could not (Figure 4F). These results demonstrated that FIT-039 suppressed the replication of a broad spectrum of DNA viruses.
Figure 4. FIT-039 suppressed the replication of HSV-2, HCMV, and HAdVs.
(A) FIT-039 suppressed the plaque formation caused by the HSV-2 infection in a dose-dependent manner. HSV-2 plaque reduction assay was performed in Vero cells treated with the indicated concentrations of FIT-039 and ACV. (B) FIT-039 suppressed the replication of HSV-2 genomic DNA in a dose-dependent manner. HSV-2 replication was analyzed by real-time PCR at the indicated concentrations of FIT-039. ACV was used as a positive control. (C) FIT-039 suppressed the replication of HCMV genomic DNA in a dose-dependent manner. HCMV replication was analyzed by real-time PCR at the indicated concentrations of FIT-039. GCV was used as a positive control. (D and E) FIT-039 suppressed the replication of HAdV genomic DNA in a dose-dependent manner. HAdV replication was analyzed by real-time PCR at the indicated concentrations of FIT-039 and CDV. (D) HAdV type 5 and (E) HAdV type 53 were examined. CDV was used as a positive control. (F) FIT-039 inhibited transcription of the HAdV gene E1A. A549 cells were infected with HAdV-5 and treated with FIT-039 at 10 μM for 6 hours. The cells were subjected to RT-PCR. In B–E, each point represents the average ± SD of the results from 3 experiments preformed in duplicate. *P < 0.001, versus DMSO treatment, Student’s t test.
Therapeutic effect of FIT-039 on mice infected with ACV-resistant HSV-1.
We next examined the antiviral effect of FIT-039 on ACV-resistant HSV-1. In the plaque reduction assay, FIT-039 significantly reduced the plaque number in Vero cells infected with ACV-resistant HSV-1, whereas GCV did not, due to cross resistance (Figure 5A). Furthermore, the effect of FIT-039 on ACV-resistant HSV-1 infection was also evaluated in the murine zosteriform spread model, in which treatment with ACV ointment did not prevent the development of lesions (Figure 5B). In contrast, treatment with the FIT-039 ointment prevented zosteriform spread (Figure 5B) and rescued mice from lethality (Figure 5C), which indicated that FIT-039 was also therapeutically effective against ACV-resistant HSV-1.
Figure 5. FIT-039 inhibited the replication of ACV-resistant HSV-1.
(A) FIT-039 suppressed the plaque formation caused by the ACV-resistant HSV-1 infection in a dose-dependent manner. Plaque reduction assay was performed in Vero cells treated with the indicated concentrations of FIT-039, ACV, and GCV. Living cells were stained with crystal violet, and the number of plaques was then counted. ACV and GCV did not suppress the plaque formation. Each point represents the average ± SD of the results from 3 experiments preformed in duplicate. *P < 0.001, versus DMSO treatment, Student’s t test. (B and C) The scratched skins of mice were infected intracutaneously with ACV-resistant HSV-1 and were then treated with ointments containing the indicated concentrations of FIT-039. Five mice were assigned to each experimental group. (B) The lesion score and (C) survival rate were determined. (B) Representative photographs of the infected mice 7 days after infection are shown. The ACV ointment did not suppress skin lesions or lethality. Each point represents the average of the results (n = 5).
Discussion
In this study, we successfully identified the novel CDK9 inhibitor FIT-039 and showed that CDK9 is a promising drug target for a broad spectrum of DNA viruses. CDK9 paired with cyclin T1 forms the human P-TEFb complex and stimulates productive transcription through phosphorylation of the CTD of RNA polymerase II and viral proteins, such as the ICP22 of HSV. The ICP22-CDK9 complex was reported previously to phosphorylate the CTD of RNA polymerase II and regulate the transcription of HSV (20, 22). A previous study showed that CDK9 was localized to nuclear viral transcriptosomes at the immediate-early stage of the HCMV infection, which allowed the active transcription of HCMV IEGs (14). The suppression of HAdV replication by FIT-039 demonstrated that HAdVs also depend on the CDK9-regulated transcription elongation machinery for their synthesis of viral RNAs. Vijayalingam et al. reported recently that the HAdV large-E1A protein activated viral transcription by recruiting the RNA polymerase II mediator complex (33); however, the effect of CDK9 on large-E1A–mediated transcription remains to be clarified. The above data suggest that these viruses take advantage of the viral protein CDK9 complexes to hijack the host transcription and perform the viral-specific transcription.
The IC50 value of FIT-039 for CDK9 inhibition is higher than the EC50 value of FIT-039 for anti–HSV-1 activity. The IC50 values of kinases show the comparative specificity of the compound but cannot be compared directly with the effect in vivo, because the kinase assay condition may not recapitulate the intracellular environments. However, we cannot deny the possibility that the antiviral activity of FIT-039 is due to a synergistic effect by inhibiting kinase(s) other than CDK9. The CTD phosphorylation is catalyzed by CDK7, CDK8, CDK11, CDK12, and CDK13, in addition to CDK9. Our preliminary experiment using siRNA against these CDKs showed that knockdown of CDK11 or CDK13 suppressed the HSV-1 replication, though the suppression was not as potent as that of CDK9 (data not shown). Furthermore, FIT-039 potently inhibited kinases, as shown in the large-panel kinase screening (see Supplemental Methods and Supplemental Table 1). GSK3 (34), PAK3 (35), DYRK3 (36), Pim1 (37), PKG1 (38), p70s6k (39), Flt3 (40), and MSK1 (41) were reported to be involved in the replication of various viruses. In addition, 6BIO, an inhibitor of GSK3, and its derivatives were reported to suppress the replication of HSV-1 and HIV (42, 43).
FIT-039 selectively inhibited viral transcription with minimal effect on cellular transcription, indicating the possibility that cellular transcription is regulated not only by CDK9 but also other CDKs, such as CDK7, CDK8, CDK11, CDK12, and CDK13, and that viral gene expression may more depend on the ICP22-CDK9 complex than cellular genes. Thus, it is conceivable that FIT-039 produced less effect on cellular transcription than on viral transcription. This study suggests that FIT-039 is a promising drug against a broad spectrum of DNA viruses.
In addition to DNA viruses, cyclin T1/CDK9 has been shown to interact with RNA viral proteins, such as the influenza virus polymerase of influenza A virus and Tat proteins of HIV, and facilitate its association with cellular RNA polymerase II (44, 45). Flavopiridol inhibited the transcription elongation activity and suppressed replication of the influenza virus (46, 47). The anti-influenza virus activity of FIT-039 was also examined. FIT-039 suppressed the replication of influenza virus H1N1 (Supplemental Figure 6). FIT-039 inhibited the HIV-Tat–dependent transcription (Supplemental Figure 7), which is regulated by CDK9 (48, 49). Furthermore, FIT-039 also suppresses the replication of HIV (M. Okamoto and M. Baba, et al., unpublished observations). These studies demonstrate the possibility that FIT-039 suppresses the replication of RNA viruses as well as DNA viruses. A drug such as FIT-039, which selectively targets the key molecules in host cells and exhibits an antiviral effect on a broad spectrum of viruses, may be able to cope with new types of viruses emerging in the future.
We are now preparing a clinical trial for FIT-039 in patients infected with ACV-resistant HSV-1 and thus investigating toxicities of FIT-039. Although mice with knockout of the target kinases, which FIT-039 potently inhibited in this study, are known to show the phenotypes raised in Supplemental Table 3, our ongoing preclinical studies have indicated that FIT-039 does not induce significant adverse effects under a high-dose oral administration protocol in rats and dogs (Supplemental Figure 8), mutagenic toxicity (Supplemental Tables 4 and 5), or irritation activity on the skin. These results are consistent with the effects of FIT-039 on cells, which does not affect cell cycle progression or cell viability.
Considering the results of toxicity tests, FIT-039 could be available for systemic treatment. However, the topical medicines described in this manuscript have following advantages: (a) the drug is easily delivered to the infectious site, and (b) the risk of adverse effects can be further reduced. Therefore, FIT-039 ointment is also unlikely to cause any side effect in the preclinical study. Furthermore, FIT-039 has an agreeable pharmacokinetic profile (Supplemental Figure 9). These results indicate that FIT-039 could be developed as oral preparations against viral infectious diseases, such as herpetic encephalitis, HCMV infectious mononucleosis, and HAdV pharyngitis. These outcomes indicate that FIT-039 is a promising novel antiviral agent for clinical therapeutics.
Methods
Viruses and cells.
The HSV strains used were HSV-1 (F strain) and HSV-2 (G strain), which were provided by Y. Kawaguchi, University of Tokyo, Tokyo, Japan. The HCMV strain used was the Towne strain, which was provided by N. Shimizu, Tokyo Medical and Dental University, Tokyo, Japan. The following HAdV strains were used: HAdV-5, obtained from National Institute of Infectious Diseases, Japan, as well as HAdV-19, HAdV-36, and HAdV-53, provided by K. Aoki and S. Ohno, Hokkaido University, Sapporo, Japan. These virus stocks were prepared in Vero cells and quantified by the plaque reduction assay. Human cervix epithelial (HeLa cells, CCL-2), African green monkey kidney (Vero cells, CCL-81), human lung fibroblast (HFL-1 cells, CCL-153), human embryonic kidney (HEK293 cells, CRL-1573), and human lung epithelial (A549 cells, CCL185) cells were grown in Minimum Essential Medium (Nacalai Tesque Inc.) supplemented with 10% FBS (Nichirei Biosciences Inc.). These cells were purchased from ATCC.
Isolation of ACV-resistant HSV-1.
ACV-resistant HSV-1 was obtained from Vero cells infected with HSV-1, which were cultured in EMEM with 1% FBS containing 20 μM ACV. Plaques appeared 2 days after the infection, and we then isolated ACV-resistant HSV from these plaques. Viral titers were determined as PFUs per ml using a plaque-forming assay in Vero cells.
Drugs.
ACV was purchased as a powder from Wako Pure Chemical Industries Ltd. GCV and CDV were purchased as powders from Sigma-Aldrich.
RNA interference.
The inhibition of CDK9 and CDK4 was achieved by siRNA Design Service (Nippon Gene Co. Ltd.) and Silencer Select siRNAs (Life Technologies Japan Ltd.). siRNA was transfected using Lipofectamine RNAiMAX (Life Technologies Japan Ltd.) according to the manufacturer’s instructions. The downregulation of CDK9 and CDK4 was investigated by Western blotting 48 hours after transfection. The siRNA sequences used for our experiments were as follows: CDK9 siRNA-1, 5′-GGAGAAUUUUACUGUGUUUdTdT-3′; CDK9 siRNA-2, 5′-CCGCUGCAAGGGUAGUAUAdTdT-3′; CDK4 siRNA, 5′-UGCUGACUUUUAACCCACAdTdT-3′; and negative control siRNA, 5′-GGUUUAGGGUAUAUUCUAUdTdT-3′. Negative control siRNA was a scramble sequence of CDK9 siRNA-1.
Antibodies and immunoblotting.
Anti-CDK9 (D-7) (Santa Cruz Biotechnology Inc.), anti-CDK4 (DCS156) (Cell Signaling Technology Inc.), anti-GAPDH (Ambion), anti-RNA polymerase II (Millipore), and goat pAb to Ms IgG (Abcam) were used in this study. Total proteins were separated in Bolt 4%–12% Bis-Tris Plus (Life Technologies), transferred to a PVDF membrane, and probed with the antibodies listed above. Signals were developed by Super Signal West Dura (Thermo Scientific) and then detected and captured by ChemiDoc (Bio-Rad).
Analysis of the cell cycle.
HeLa cells were plated in a 6-cm culture dish at a density of 1 × 105 cells per dish. Cells were treated with siRNAs for 48 hours or incubated with FIT-039 or flavopiridol at 4 μM or 800 nM, respectively. Cells were stained with propidium iodide following these incubations and analyzed using FACSCalibur (BD Biosciences).
CDK9 inhibitor screening.
We screened our kinase-focused library consisted of 144 compounds. In vitro kinase assays were performed in 96-well format using 100 ng CDK9 (Carna Biosciences), 2 μM CDK7/9 tide (Millipore), and 100 nM ATP per well in reaction buffer (8 mM of MOPS-NaOH [pH 7.0] and 20 μM EDTA) with 1 μM compounds or 4% DMSO as negative control. After 2 hours of incubation at 30°C, the amounts of remaining ATP in the kinase reaction were quantified by kinase Glo (Promega). The inhibition of kinase activity by each compound was calculated as follows: inhibition (%) = [1 – (A – B)/(C – B)] × 100, where A is the response with the compound, B is the background response with kinase, and C is the response with negative control (4% DMSO).
Docking simulation study.
Three crystal structures of human CDK9 (PDB IDs: 3TN8, 3BLQ, 3BLR) were selected as the template for docking simulations with FIT-039. Simulations were performed by CDOCKER and related modules in Discovery Studio 3.0 (Accelrys Inc. USA). The same procedure was applied to human CDK4 (PDB IDs: 2W9Z, 2W96) but failed to produce a reasonable complex model. Thus, the CDK4 structure was superimposed on that of CDK9 in the CDK9/FIT-039 complex.
Plaque reduction assay.
Wild-type and ACV-resistant HSV-infected Vero cells were incubated with 150 PFUs per 1 ml for 1 hour at 37°C, and the culture media were then changed to fresh media containing various concentrations of FIT-039 and ACV. After 2 days of incubation, the infected cells were fixed with 100% methanol and stained with crystal violet solution (2.5% crystal violet, 30% ethanol, and 1% ammonium oxalate). The number of plaques was counted under a microscope.
Quantitative PCR analysis.
HeLa cells, HFL-1 cells, and A549 cells were infected with HSVs, HCMV, and HAdVs, respectively, at a MOI of 0.1, with chemical compounds. HSV-, HCMV-, and HAdV-infected cells were incubated for 24, 72, and 24 hours, respectively. After incubation, these cells were harvested and their DNA was isolated. HSVs, HCMV, and HAdVs, genomic DNA, and cellular GAPDH were quantitative by real-time PCR. Analyses of HSV-1, HSV-2, HAdV-5, and GAPDH were performed using FastStart Universal Probe Master (ROX) (Roche Applied Science). Analysis of HAdV-19 was performed using FastStart Universal SYBR Green Master (Roche Applied Science). PCR was performed with an initial denaturation reaction at 95κC for 1 minute and then amplified with 40 cycles of 95°C for 30 seconds, 60°C for 30 seconds, and 72°C for 30 seconds. The amplification was monitored on Step One Plus (Applied Biosystems Inc.). The primers used are shown in Supplemental Table 5.
MTT assay.
Cytotoxicity was evaluated by the MTT reduction assay. HeLa cells were seeded at a concentration of 1 × 104 cells per well in 96-well tissue culture plates and grown at 37°C for 1 day. The culture medium was replaced by fresh medium containing FIT-039 at various concentrations (1–50,000 nM), and cells were grown for a further 24 hours. After incubation, the water-soluble tetrazolium salt, WST-8 (Nacalai Tesque Inc.), solution was added to media. Cells were incubated at 37°C for 2 hours, followed by plate reading on a ARVO X5 microplate reader (PerkinElmer Inc.), using a test wavelength of 450 nm and reference wavelength of 600 nm.
RT-PCR and quantitative RT-PCR analysis.
HeLa cells and A549 cells were infected with HSV-1 and HAdV-5, respectively, at MOI of 0.1, with chemical compounds. HSV-1– and HAdV-5–infected cells were incubated for 3 hours or 6 hours, respectively, following total RNA extraction using Sepasol RNA-I Super (Nacalai Tesque Inc.). Reverse transcription was performed with PrimeScript Reverse Transcriptase (Takara Bio Inc.), using oligo (dT) primer. The IEGs of HSV-1, E1A of HAdV, and cellular GAPDH were then amplified by PCR. PCR was performed using PrimeStar HS DNA Polymerase with GC Buffer (Takara Bio Inc.). PCR was performed with an initial denaturation reaction at 95°C for 1 minute and then amplified with 27 cycles of 98°C for 10 seconds, 60°C for 5 seconds, 72°C for 1 minute. This amplification was monitored on a T100 Thermal Cycler (Bio-Rad). The primers used are shown in Supplemental Tables 6 and 7.
Microarray analysis.
HeLa cells were plated in a 6-cm culture dish at a density of 1 × 105 cells per dish. Cells were incubated with FIT-039 or flavopiridol at 4 μM or 800 nM, respectively, following total RNA extraction using the RNeasy Mini Kit (QIAGEN), per the manufacturer’s instructions. RNA was prepared for microarray expression analysis by the GeneChip 3′ IVT Expression Kit (Affymetrix Inc.) according to the manufacturer’s instructions. The prepared RNA was hybridized and stained onto the Human Genome U133 Plus 2.0 array (Affymetrix Inc.) using the GeneChip Hybridization Wash and Stain Kit (Affymetrix Inc.). Signals were detected using the GeneChip Scanner 3000G (Affymetrix Inc.). These data were analyzed with Perl scripts and a statistical package, R (http://www.r-project.org/).
Two-week repeat-dose oral toxicity study.
Male 5-week-old Wister rats were purchased from Charles River Laboratories Inc. FIT-039 (1,000 mg/kg) or the solvent (carboxymethylcellulose) was administrated orally to male rats once a day for 14 days, and these animals were followed for 14 days from the final administration. Three rats were assigned to each experimental group. Body weights and general conditions were determined for 28 days from the first administration. Hematology tests were performed and the organ weights were noted on day 28.
Ointment supplement with chemical compounds.
The 5% ACV ointment, which contained polyethylene glycol, glycerol, and ACV, was provided by Rakool Pharmaceutical Marketing Co. Ltd. Japan. We used 10% lactic acid, in addition to polyethylene glycol and glycerol, to dissolve FIT-039 in ointment.
Mouse skin HSV-1 or ACV-resistant HSV-1 infection model.
Male 5-week-old ICR mice were purchased from Charles River Laboratories Inc. The left mid-flank of each mouse was depilated with a chemical depilatory hair remover. The naked skin was scratched 1 or 2 days later with a 27-gauge needle, and 20 μl of the HSV-1 suspension of 2 × 105 PFUs was applied to the scratched area. Each ointment was administered to mice (5 mice per group) as follows. For experiment 1 (topical treatment, the effect of different concentrations), FIT-039 prepared in the ointment formulation at concentrations of 1.25%, 2.5%, 5%, and 10% (w/w) was topically applied to the scratched area (30 mg/area/dose) 3 hours after the HSV-1 infection and twice daily for 12 days. For experiment 2 (topical treatment, therapeutic efficacy of FIT-039 in skin ACV-resistant HSV-1–infected mice), 10% FIT-039 ointment was applied topically to the scratched area (30 mg/area/dose) 3 hours after the ACV-resistant HSV-1 infection and twice daily for 12 days. The placebo (ointment base) and 5% ACV ointment applied twice daily were used as control. In both experiments, the development of skin lesions and mortality were continuously observed twice daily for 12 days after the HSV-1 or ACV-resistant HSV-1 infection and scored as follows: 0, no lesion; 2, vesicles in a local region; 4, erosion and/or ulceration in a local region; 6, mild zosteriform lesion; 8, moderate zosteriform lesion; 10, severe zosteriform lesion; 14, death.
Database repository.
Gene expression profiling data were deposited in GEO (accession no. GSE57429).
Statistics.
Numerical values are reported as mean ± SD unless stated otherwise. Statistical significance was determined with a 2-tailed Student’s t test. A P value of less than 0.05 was considered significant.
Study approval.
All animal protocols were reviewed and approved by the Animal Research Committee, Graduate School of Medicine, Kyoto University.
Supplementary Material
Acknowledgments
We thank Kei Aoyama, Chikako Murasaki, and Takako Koike (Department of Biological Information, Graduate School of Bioscience and Biotechnology, Tokyo Institute of Technology) for their contributions on the synthesis of small compounds; Ayako Hosoya (Tokyo Medical and Dental University) for HRMS analyses; Koki Aoki and Shigeaki Ohno (Department of Ophthalmology, Graduate School of Medicine, Hokkaido University) for ADV types 19, 37, and 53; Youich Kobayashi, Minoru Morita, Yuushin Akimoto, Shigeru Imoto, Yukihiro Shibuya, and Takuya Kato (Rakool Pharmaceutical Marketing Co. Ltd. Japan) for the FIT-039 ointment; Kenji Ooe for help writing the manuscript; and Yasushi Kawaguchi, Daisuke Watanabe, and our laboratory members for their critical comments and discussion. We also thank the Institute of Laboratory Animals Graduate School of Medicine and the Radioisotope Research Center of Kyoto University for their instrumental support. This work was supported by Grants-in-Aid for Scientific Research (21249013 to M. Hagiwara), Research Program of Innovative Cell Biology (231006 to M. Hagiwara), and Platform for Drug Discovery (to M. Hagiwara) from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan and by grants from National Institute of Biomedical Innovation (241011 to M. Hagiwara), CREST (231038 to M. Hagiwara), and Risk-Taking Fund for Technology Development (D06-14 to KinoPharma Inc.) from Japan Science and Agency.
Footnotes
Conflict of interest: Makoto Yamamoto and Hiroshi Onogi are employees of KinoPharma Inc. Masatoshi Hagiwara owns equity in and is a consultant for KinoPharma Inc.
Reference information:J Clin Invest. 2014;124(8):3479–3488. doi:10.1172/JCI73805.
References
- 1.Ostertag W, et al. Induction of endogenous virus and of thymidine kinase by bromodeoxyuridine in cell cultures transformed by Friend virus. Proc Natl Acad Sci U S A. 1974;71(12):4980–4985. doi: 10.1073/pnas.71.12.4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elion GB, Furman PA, Fyfe JA, de Miranda P, Beauchamp L, Schaeffer HJ. Selectivity of action of an antiherpetic agent, 9-(2-hydroxyethoxymethyl) guanine. Proc Natl Acad Sci U S A. 1977;74(12):5716–5720. doi: 10.1073/pnas.74.12.5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sellar RS, Peggs KS. Management of multidrug-resistant viruses in the immunocompromised host. Br J Haematol. 2012;156(5):559–572. doi: 10.1111/j.1365-2141.2011.08988.x. [DOI] [PubMed] [Google Scholar]
- 4.Field HJ. Herpes simplex virus antiviral drug resistance--current trends and future prospects. J Clin Virol. 2001;21(3):261–269. doi: 10.1016/S1386-6532(00)00169-4. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y, Wang Q, Zhu Q, Zhou R, Liu J, Peng T. Identification and characterization of acyclovir-resistant clinical HSV-1 isolates from children. J Clin Virol. 2011;52(2):107–112. doi: 10.1016/j.jcv.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 6.Karlas A, et al. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature. 2010;463(7282):818–822. doi: 10.1038/nature08760. [DOI] [PubMed] [Google Scholar]
- 7.Schang LM. Cyclin-dependent kinases as cellular targets for antiviral drugs. J Antimicrob Chemother. 2002;50(6):779–792. doi: 10.1093/jac/dkf227. [DOI] [PubMed] [Google Scholar]
- 8.Siakallis G, Spandidos DA, Sourvinos G. Herpesviridae and novel inhibitors. Antivir Ther. 2009;14(8):1051–1064. doi: 10.3851/IMP1467. [DOI] [PubMed] [Google Scholar]
- 9.Kusano-Kitazume A, et al. Identification of novel N-(morpholine-4-carbonyloxy) amidine compounds as potent inhibitors against hepatitis C virus replication. Antimicrob Agents Chemother. 2012;56(3):1315–1323. doi: 10.1128/AAC.05764-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anwar A, et al. The kinase inhibitor SFV785 dislocates dengue virus envelope protein from the replication complex and blocks virus assembly. Plos One. 2011;6(8):e23246. doi: 10.1371/journal.pone.0023246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karakama Y, et al. Inhibition of hepatitis C virus replication by a specific inhibitor of serine-arginine-rich protein kinase. Antimicrob Agents Chemother. 2010;54(8):3179–3186. doi: 10.1128/AAC.00113-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fukuhara T, et al. Utilization of host SR protein kinases and RNA-splicing machinery during viral replication. Proc Natl Acad Sci U S A. 2006;103(30):11329–11333. doi: 10.1073/pnas.0604616103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Duyne R, et al. Effect of mimetic CDK9 inhibitors on HIV-1-activated transcription. J Mol Biol. 2013;425(4):812–829. doi: 10.1016/j.jmb.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kapasi AJ, Clark CL, Tran K, Spector DH. Recruitment of cdk9 to the immediate-early viral transcriptosomes during human cytomegalovirus infection requires efficient binding to cyclin T1, a threshold level of IE2 86, and active transcription. J Virol. 2009;83(11):5904–5917. doi: 10.1128/JVI.02651-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schang LM, St Vincent MR, Lacasse JJ. Five years of progress on cyclin-dependent kinases and other cellular proteins as potential targets for antiviral drugs. Antivir Chem Chemother. 2006;17(6):293–320. doi: 10.1177/095632020601700601. [DOI] [PubMed] [Google Scholar]
- 16.Tan SL, Ganji G, Paeper B, Proll S, Katze MG. Systems biology and the host response to viral infection. Nat Biotechnol. 2007;25(12):1383–1389. doi: 10.1038/nbt1207-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9(3):153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 18.Kohoutek J, Blazek D. Cyclin K goes with Cdk12 and Cdk13. Cell Div. 2012;7:12. doi: 10.1186/1747-1028-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evers DL, Breitenbach JM, Borysko KZ, Townsend LB, Drach JC. Inhibition of cyclin-dependent kinase 1 by purines and pyrrolo[2,3-d]pyrimidines does not correlate with antiviral activity. Antimicrob Agents Chemother. 2002;46(8):2470–2476. doi: 10.1128/AAC.46.8.2470-2476.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feichtinger S, et al. Recruitment of cyclin-dependent kinase 9 to nuclear compartments during cytomegalovirus late replication: importance of an interaction between viral pUL69 and cyclin T1. J Gen Virol. 2011;92(pt 7):1519–1531. doi: 10.1099/vir.0.030494-0. [DOI] [PubMed] [Google Scholar]
- 21.Rechter S, et al. Cyclin-dependent kinases phosphorylate the cytomegalovirus RNA export protein pUL69 and modulate its nuclear localization and activity. J Biol Chem. 2009;284(13):8605–8613. doi: 10.1074/jbc.M805693200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Durand LO, Roizman B. Role of cdk9 in the optimization of expression of the genes regulated by ICP22 of herpes simplex virus 1. J Virol. 2008;82(21):10591–10599. doi: 10.1128/JVI.01242-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Biglione S, et al. Inhibition of HIV-1 replication by P-TEFb inhibitors DRB, seliciclib and flavopiridol correlates with release of free P-TEFb from the large, inactive form of the complex. Retrovirology. 2007;4:47. doi: 10.1186/1742-4690-4-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ou M, Sandri-Goldin RM. Inhibition of cdk9 during herpes simplex virus 1 infection impedes viral transcription. PLoS One. 2013;8(10):e79007. doi: 10.1371/journal.pone.0079007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meinhart A, Kamenski T, Hoeppner S, Baumli S, Cramer P. A structural perspective of CTD function. Genes Dev. 2005;19(12):1401–1415. doi: 10.1101/gad.1318105. [DOI] [PubMed] [Google Scholar]
- 26.Sanchez V, et al. Cyclin-dependent kinase activity is required at early times for accurate processing and accumulation of the human cytomegalovirus UL122-123 and UL37 immediate-early transcripts and at later times for virus production. J Virol. 2004;78(20):11219–11232. doi: 10.1128/JVI.78.20.11219-11232.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schang LM, et al. Pharmacological cyclin-dependent kinase inhibitors inhibit replication of wild-type and drug-resistant strains of herpes simplex virus and human immunodeficiency virus type 1 by targeting cellular, not viral, proteins. J Virol. 2002;76(15):7874–7882. doi: 10.1128/JVI.76.15.7874-7882.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delehouze C, et al. CDK/CK1 inhibitors roscovitine and CR8 downregulate amplified MYCN in neuroblastoma cells. Oncogene. doi: 10.1038/onc.2013.513. [published online ahead of print December 9, 2013]. doi:10.1038/onc.2013.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas JP, et al. Phase I clinical and pharmacokinetic trial of the cyclin-dependent kinase inhibitor flavopiridol. Cancer Chemother Pharmacol. 2002;50(6):465–472. doi: 10.1007/s00280-002-0527-2. [DOI] [PubMed] [Google Scholar]
- 30.Diwan P, Lacasse JJ, Schang LM. Roscovitine inhibits activation of promoters in herpes simplex virus type 1 genomes independently of promoter-specific factors. J Virol. 2004;78(17):9352–9365. doi: 10.1128/JVI.78.17.9352-9365.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simmons A, Nash AA. Zosteriform spread of herpes simplex virus as a model of recrudescence and its use to investigate the role of immune cells in prevention of recurrent disease. J Virol. 1984;52(3):816–821. doi: 10.1128/jvi.52.3.816-821.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Betz UA, Fischer R, Kleymann G, Hendrix M, Rubsamen-Waigmann H. Potent in vivo antiviral activity of the herpes simplex virus primase-helicase inhibitor BAY 57-1293. Antimicrob Agents Chemother. 2002;46(6):1766–1772. doi: 10.1128/AAC.46.6.1766-1772.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vijayalingam S, Chinnadurai G. Adenovirus L-E1A activates transcription through mediator complex-dependent recruitment of the super elongation complex. J Virol. 2013;87(6):3425–3434. doi: 10.1128/JVI.03046-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Konig R, et al. Human host factors required for influenza virus replication. Nature. 2010;463(7282):813–817. doi: 10.1038/nature08699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nguyen DG, Wolff KC, Yin H, Caldwell JS, Kuhen KL. “UnPAKing” human immunodeficiency virus (HIV) replication: using small interfering RNA screening to identify novel cofactors and elucidate the role of group I PAKs in HIV infection. J Virol. 2006;80(1):130–137. doi: 10.1128/JVI.80.1.130-137.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bakre A, et al. Identification of host kinase genes required for influenza virus replication and the regulatory role of microRNAs. PLoS One. 2013;8(6):e66796. doi: 10.1371/journal.pone.0066796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng F, et al. KSHV reactivation from latency requires Pim-1 and Pim-3 kinases to inactivate the latency-associated nuclear antigen LANA. PLoS Pathog. 2009;5(3):e1000324. doi: 10.1371/journal.ppat.1000324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee JH, Yedavalli VR, Jeang KT. Activation of HIV-1 expression and replication by cGMP dependent protein kinase type 1-β (PKG1β). Retrovirology. 2007;4:91. doi: 10.1186/1742-4690-4-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coito C, Diamond DL, Neddermann P, Korth MJ, Katze MG. High-throughput screening of the yeast kinome: identification of human serine/threonine protein kinases that phosphorylate the hepatitis C virus NS5A protein. J Virol. 2004;78(7):3502–3513. doi: 10.1128/JVI.78.7.3502-3513.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arita M, Wakita T, Shimizu H. Cellular kinase inhibitors that suppress enterovirus replication have a conserved target in viral protein 3A similar to that of enviroxime. J Gen Virol. 2009;90(pt 8):1869–1879. doi: 10.1099/vir.0.012096-0. [DOI] [PubMed] [Google Scholar]
- 41.Jamaluddin M, Tian B, Boldogh I, Garofalo RP, Brasier AR. Respiratory syncytial virus infection induces a reactive oxygen species-MSK1-phospho-Ser-276 RelA pathway required for cytokine expression. J Virol. 2009;83(20):10605–10615. doi: 10.1128/JVI.01090-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hsu MJ, Hung SL. Antiherpetic potential of 6-bromoindirubin-3′-acetoxime (BIO-acetoxime) in human oral epithelial cells. Arch Virol. 2013;158(6):1287–1296. doi: 10.1007/s00705-013-1629-3. [DOI] [PubMed] [Google Scholar]
- 43.Guendel I, et al. Novel neuroprotective GSK-3β inhibitor restricts Tat-mediated HIV-1 replication. J Virol. 2014;88(2):1189–1208. doi: 10.1128/JVI.01940-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang J, Li G, Ye X. Cyclin T1/CDK9 interacts with influenza A virus polymerase and facilitates its association with cellular RNA polymerase II. J Virol. 2010;84(24):12619–12627. doi: 10.1128/JVI.01696-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bieniasz PD, Grdina TA, Bogerd HP, Cullen BR. Highly divergent lentiviral Tat proteins activate viral gene expression by a common mechanism. Mol Cell Biol. 1999;19(7):4592–4599. doi: 10.1128/mcb.19.7.4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang S, Zhang J, Ye X. [Protein kinase inhibitor flavopiridol inhibits the replication of influenza virus in vitro]. Wei Sheng Wu Xue Bao. 2012;52(9):1137–1142. [PubMed] [Google Scholar]
- 47.Ali A, et al. Identification of flavopiridol analogues that selectively inhibit positive transcription elongation factor (P-TEFb) and block HIV-1 replication. Chembiochem. 2009;10(12):2072–2080. doi: 10.1002/cbic.200900303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mancebo HS, et al. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 1997;11(20):2633–2644. doi: 10.1101/gad.11.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu Y, et al. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 1997;11(20):2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.