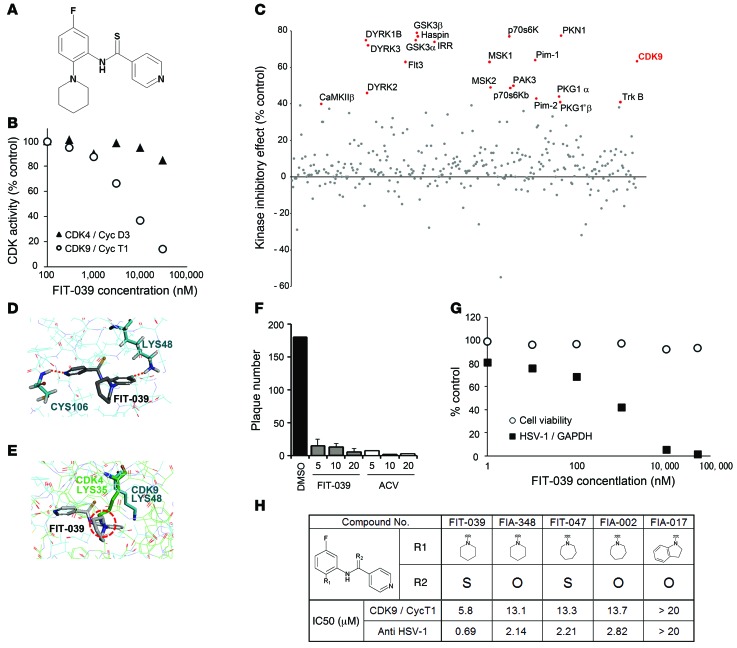
Figure 1. Identification of the CDK9-selective inhibitor FIT-039, which suppressed the replication of HSV-1.
(A) Chemical structure of FIT-039. (B) In vitro kinase assay of CDK9/cyclin T1 and CDK4/cyclinD3. An increased amount of FIT-039 inhibited CDK9/cyclin T1 but not CDK4/cyclinD3. (C) Scatter plots of the 332-kinase panel of FIT-039 (10 μM). Red dots indicate greater than 40% inhibition. (D) A structural model for FIT-039 bound to CDK9. FIT-039 formed 2 hydrogen bonds in the ATP-binding pocket, in which LYS48 and CYS106 interacted with the fluoride moiety and nitrogen atom of the pyridine of FIT-039, respectively. (E) The ATP-binding pocket of CDK4 (thin green lines) was superimposed onto that of CDK9 (thin blue lines) in the CDK9/FIT-039 complex model shown in Figure 2D. The different orientation of the side chain of LYS35 of CDK4, which corresponds to the LYS48 of CDK9, would hinder the binding of FIT-039 to CDK4. (F) FIT-039 suppressed the plaque formation caused by the HSV-1 infection in a dose-dependent manner. HSV-1 plaque reduction assay was performed in Vero cells treated with DMSO, FIT-039, and ACV. (G) FIT-039 inhibited the replication of HSV-1 genomic DNA in a dose-dependent manner without cytotoxicity. HSV-1 replication and cytotoxicity were analyzed by real-time PCR and the WST-8 assay, respectively. (H) Structure-activity relationship. The chemical structures of FIT-039 and its derivatives are shown. IC50 values of anti–HSV-1 activity were determined by real-time PCR of HSV-1 genomic DNA replication in infected HeLa cells. The IC50 values of CDK9/cyclin T1 inhibitory activity were calculated by the in vitro kinase assay. The IC50 values of anti–HSV-1 activity were correlated with those of CDK9/cyclin T1 activity (R2 = 0.92).