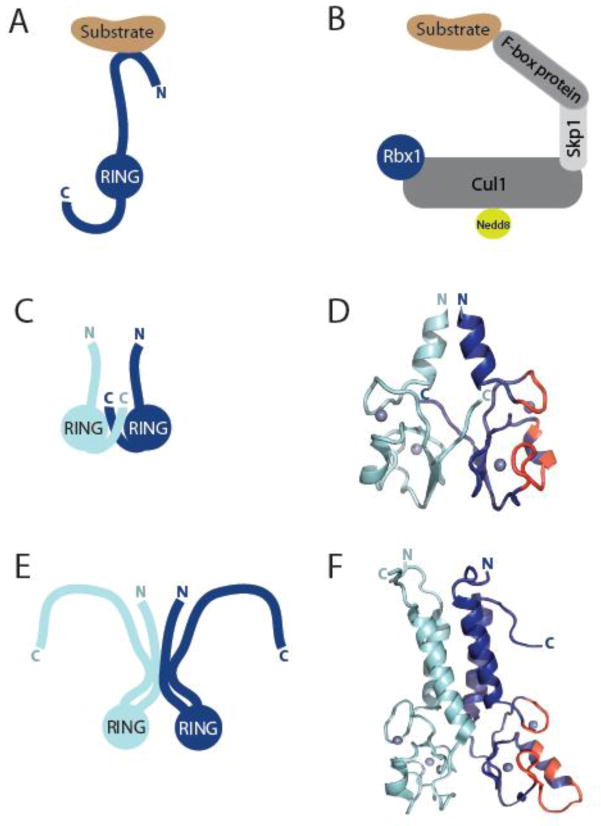
Figure 3. Architecture of RING-type E3s.
A) Model of a monomeric RING-type E3, where its RING domain would mediate binding to E2 thioester-linked to ubiquitin. Binding to substrate occurs generally through regions of the E3 other than the RING domain. B) Model of a multi-subunit RING E3 of the Cullin RING Ligase (CRL) superfamily, such as the well-studied SCF (CRL1) family, shown here. SCF consists of a cullin protein (Cul1) a small RING finger protein (Rbx1), and an adaptor protein (Skp1) that binds interchangeable substrate recognition elements (F-box proteins). The ubiquitin-like molecule, Nedd8, is reversibly conjugated to cullins and associated with activation of CRLs. C) Schematic of dimeric RING E3s, such as cIAP, RNF4, BIRC7, IDOL, Mdm2-MdmX, that dimerize through their RING domains and interleaved C-terminal tails. D) Ribbon diagram illustrating the homodimeric RING E3, BIRC7 [13] (PDB 4AUQ) as a representative of the class of dimers schematized in C. The E2-interacting residues of one RING domain are highlighted in red. E) Model of dimeric RING E3s, such as BRCA1-BARD1 and RING1B-Bmi1, where α-helices both N- and C-terminal to the RING facilitate dimerization. In the case of BRCA1-BARD1 (illustrated), this occurs through a four α-helix bundle (helices above RINGs in F). F) Ribbon diagram illustrating the heterodimeric RING dimer of BRCA1-BARD1 [19] (PDB 1JM7) modeled in E). The E2-interacting residues of the RING domain of BRCA1 are highlighted in red.