Figure 4. Activation of E2~Ub conjugates by RING-type domains.
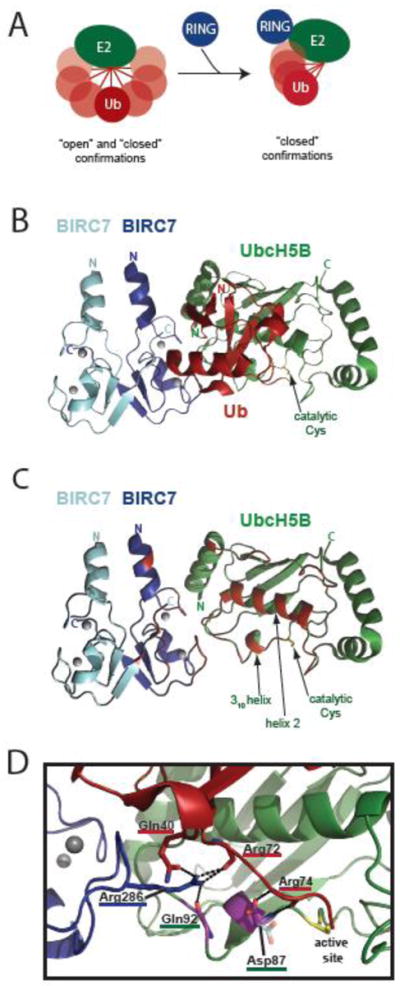
A) Schematic of ubiquitin thioester linked to E2 sampling ‘open’ and ‘closed’ confirmations in the absence of a RING domain (left). RING binding to E2~Ub promotes increased occupancy of closed confirmations needed for ubiquitin transfer (right). B) Ribbon diagram of BIRC7-BIRC7:UbcH5B~Ub (PDB 4AUQ) as an example of a RING-type E3:E2~Ub ternary complex. E3, E2, and ubiquitin are blue, green, and red, respectively. The catalytic Cys of UbcH5B is labeled. Gray circles are the zincs coordinated by the BIRC7 RINGs. C) The ribbon diagram shown in B, with the ubiquitin molecule removed and the residues on E2 or the RING domains that would contact ubiquitin in red. The 310 helix, helix 2, and catalytic Cys of UbcH5B are labeled. D) A closer view of the E2 active site and ubiquitin C-terminus from the BIRC7-BIRC7:UbcH5B~Ub structure shown in B and C. The UbcH5B helix 2 (green) is positioned directly above Arg72. Residues from BIRC7, UbcH5B, and ubiquitin are underlined in blue, green, and red respectively. The hydrogen bonding network created by BIRC7 Arg286 (blue) and the E2 backbone of Gln92, the ubiquitin backbone of Arg72, and the side chain of ubiquitin Gln40 is shown. Residues colored in purple correspond to those that display NMR spectral effects specifically arising from the E3:E2 hydrogen bond. The ‘up’ conformation of the Asp87 side chain seen in this structure is shown in purple and red, the ‘down’ conformation, frequently seen in structures in the absence of covalently-bound ubiquitin is shown coming off the backbone directly below in semi-transparent cyan and orange (taken from PDB 3UGB [170]). Contacts made by the ‘up’ Asp87 conformation to Arg74 (red) of ubiquitin are shown. The side chains of UbcH5B Gln92, Ub Arg72, and Ub Arg74 are not shown for clarity.
