Abstract
Over-expression of the oncoprotein, Aurora A kinase occurs in multiple types of carcinomas, often early during cell transformation. To identify mechanism(s) contributing to enhanced Aurora A protein expression, we examined normal human lung fibroblast and breast epithelial cells and compared them to non-tumorigenic breast (MCF10A and MCF12A) and tumorigenic breast and cervical epithelial cell lines (MCF-7 and HeLa S3, respectively). A subset of these immortalized lines (MCF10, MCF12A, and HeLa S3) exhibited increased levels of Aurora A protein, independent of tumorigenicity. The increase in Aurora A protein expression in these immortalized cells was not due to increased transcription/RNA stability, protein half-life or cap-dependent translation. Assays utilizing monocistronic and dicistronic RNA constructs revealed that the Aurora A 5′ leader contains an internal ribosomal entry site (IRES), which is regulated through the cell cycle, peaking in G2/M phase. Moreover, IRES activity was increased in the immortalized cell lines in which Aurora A protein expression was also enhanced. Additional assays indicated that the increased internal initiation is specific to the Aurora A IRES and may be an early event during cancer progression. Taken together, these results identify a novel mechanism contributing to Aurora A kinase over-expression and possibly to immortalization leading to carcinogenesis.
Keywords: Protein synthesis, Internal ribosomal entry site, Aurora A, Immortalization, Breast cancer
INTRODUCTION
Aurora A is a serine/threonine kinase that plays a crucial regulatory role during mitotic events including centrosome duplication, separation, and maturation as well as mitotic spindle stabilization (1–3). Regulation of Aurora A expression is tightly controlled with both the mRNA and protein detected in late S early G2 phase, peaking in G2/M, and rapidly degrading prior to G1 phase (4,5). Aberrant expression of this kinase is detrimental to the cell. Over-expression contributes to centrosome amplification and a failure in cytokinesis creating aneuploidy, setting the stage for carcinogenesis (6,7). Numerous tumor cell lines and human tumors exhibit elevated levels of the Aurora A kinase, suggesting it may play a role in tumorigenesis (8–11). Indeed, ectopic Aurora A over-expression leads to cell transformation (12). Alternatively, loss of Aurora A leads to centrosomal separation defects resulting in a monopolar spindle, which in turn activates the G2/M checkpoint and eventually apoptosis (13). For this reason the Aurora A kinase is considered a target for the development of anticancer drugs.
Enhanced expression of Aurora A protein in tumors is reportedly due to a concomitant increase in Aurora A mRNA owing to gene amplification and/or increased transcription (7,8,14–16). However, there are examples in many cancers whereby increased Aurora A protein expression is not accompanied by changes in mRNA levels (9,10,17). These results suggest that post-transcriptional processes including enhanced protein synthesis and/or protein stability are also contributing to the increased Aurora A kinase levels.
The major regulatory step in protein synthesis occurs at the initiation of translation (18). Most eukaryotic mRNAs are thought to initiate translation in a cap-dependent manner. This mechanism involves the binding of the pre-initiation complex to the methyl-7-guansine (m7G) cap structure at the 5′ end of the mRNA and scanning of the 40S ribosome to the first initiator codon in a proper context (19) (reviewed in (20,21)). In a subset of cellular mRNAs, an alternative mechanism to initiate translation occurs in which the pre-initiation complex internally binds the 5′ leader or untranslated region (UTR) (22,23). The binding site is referred to as an internal ribosome entry site (IRES). During periods in which cap-dependent translation is decreased, including in response to cellular stress or throughout the G2/M phase of the cell cycle, IRES-dependent translation is proposed to be maintained or elevated (24–27). Indeed, many mRNAs translated during mitosis contain IRESes. For example, IRES-dependent translation of the ornithine decarboxylase (26) and PITSRLE p58 (28) mRNA occurs exclusively during mitosis (25). Additionally, most of the small subset of eukaryotic IRESes identified to date are located in mRNAs that encode proteins that affect tumorigenesis. These proteins include oncogenes (c-myc) (29), growth factors (FGF2) (30–32), growth factor receptors (TrkB) (33), pro- and anti-apoptotic factors (XIAP and APAF-1, respectively) (34,35), and angiogenic factors (VEGF) (32). Deregulating IRES-dependent translation of these mRNAs could be a mechanism to promote cell survival and uncontrolled cellular proliferation during carcinogenesis.
In the present report, we chose multiple cell lines that differentially express Aurora A protein to identify mechanism(s) contributing to its over-expression. Transcription, mRNA stability, cap-dependent translation and protein stability could not account for the increased Aurora A protein expression in a subset of cell lines. However, an IRES was identified in the 5′ leader of the Aurora A mRNA. Aurora A IRES activity positively correlated with Aurora A protein levels. Moreover, Aurora A expression in these cells was unaffected when cap-dependent translation was reduced. We propose there is a switch from cap-dependent to IRES-dependent translation of the Aurora A mRNA that contributes to over-expression of the protein. In turn, this enhanced IRES activity may be a key determinant in generating genomic instability that may eventually result in cellular immortalization.
MATERIAL AND METHODS
Constructs
The Aurora A (Supplemental Figure 1) and β-globin (GenBank: V00497.1) 5′ leaders were PCR amplified from a human fetal brain cDNA library (Clontech) and inserted into the dual luciferase vector-RP (36,37) (a generous gift from Dr Anne Willis, University of Leicester) with EcoRI and NcoI endonuclease restriction sites The dicistronic construct for in vitro transcription was created by digesting the RP vector with EcoRV and BamHI releasing the Renilla and Photinus luciferase genes and the SV40 3′-untranslated region (3′-UTR). The two luciferase genes were inserted into the multiple cloning site of the SK+ Bluescript vector (Stratagene) downstream of the T7 promoter. The monocistronic construct for in vitro transcription was created by digesting the RP vector with EcoRI and BamHI. The digest released the 5′ leader, the Photinus luciferase gene and the SV40 3′-UTR, which were inserted into the EcoRI and BamHI sites of the SK+ Bluescript vector (Stratagene) downstream of a T7 promoter.
In experiments using a hypophosphorylated form of 4E-BP1 (containing Thr-37-Ala/Thr-46-Ala/Ser-65-Ala/Thr-70-Ala/Ser-83-Ala mutations), HeLa cells were transfected with 4μg of a plasmid expressing hypophosphorylated 4E-BP1 or a control vector (Paltag) using Fugene® 6 transfection reagent (Roche). After 48 hrs, expression was analyzed via Western blotting. The 4E-BP1 mutant was generously provided by Dr. Davide Ruggero (University of California, San Francisco).
In vitro transcription
The dicistronic and monocistronic SK+ Bluescript vectors were linearized with BamHI and used as templates for in vitro transcription. For the in vitro translation assay, monocistronic templates were transcribed using mMessage mMachine® T7 Ultra (Ambion) producing capped mRNA. For the RNA transfection assays, dicistronic and monocistronic templates were transcribed using MEGAScript® T7 (Ambion) producing either A capped (New England Biolabs) or uncapped RNA. The m7G cap was added using Script Cap™ m7G capping system (Cellscript.Inc) and transcripts were poly (A) tailed using Poly(A) Polymerase tailing kit (Epicentre) per manufacturer’s instructions. All mRNA was extracted with phenol/chloroform and run on an agarose gel to ensure RNA integrity.
In vitro translation
An aliquot of 0.5 μg of in vitro transcribed mRNA, cap analog (Ambion) and 1.6 nM methionine was added to rabbit reticulocyte lysate (Speed Read, Novagen) and incubated for 1 h at 30°C. The sample was subsequently assayed for Photinus and Renilla luciferase activity.
siRNA plasmid transfections
10 mM of nonsense siRNA of (Dharmacon, D-001206-10-20) or human eIF-4E siRNA (Dharmacon, M-003884-03, J-003884-08, or J-003884-10)) were incubated in 35-mm plate wells with 12 μl of INTERFERin® transfection reagent (PolyPlus-Transfection) at 37°C for 10 min. HeLa cells were plated at 1.0×106 in wells already containing siRNA complexes and serum-free growth media. Complete growth media was then added to a final volume of 2 ml. After 48 hr, cells were harvested in cell lysis buffer (Promega) with protease (Roche) and phosphatase inhibitors (Pierce) and analyzed via Western blot.
Cell culture/Luciferase assays
WI-38 cells (CCL-75) were obtained by American Type Culture Collection (ATCC, Manassas, VA) and cultured in MEM plus 10% FBS and 5% Pen/Strep. MCF-7 (HTB-22), HeLa (CCL-2) and HeLa S3 cells (CCL-2.2) were also obtained by ATCC and cultured in DMEM plus 10% FBS and 5% Pen/Strep. HMEC cells (A10565) were obtained from Invitrogen and cultured in the recommended medium. HMEC-t cells were generously provided by Dr. James DeGregori (University of Colorado Anschutz Medical Center) and cultured in HMEC medium. MCF10A, MCF12A, and the 21 T series were generously provided by Dr. Heide Ford (University of Colorado Anschutz Medical Center). MCF10A and MCF12A were cultured as previously described (Ford 1998). 21PT, 21NT, and 21 MT2 were cultured as previously described (Schedin 2004). Cell lines were validated by STR DNA fingerprinting using the AmpFℓSTR Identifiler kit according to manufacturer instructions (Applied Biosystems cat 4322288). The STR profiles were compared to known ATCC fingerprints (ATCC.org), to the Cell Line Integrated Molecular Authentication database (CLIMA) version 0.1.200808 (http://bioinformatics.istge.it/clima/) (Nucleic Acids Research 37:D925-D932 PMCID: PMC2686526) and to the MD Anderson fingerprint database. The STR profiles matched known DNA fingerprints or were unique.
Cells were transfected with 2ug of mRNA using TransMessenger™ transfect kit (Qiagen –RNA/TransMessenger lipid 1:8) per manufacturer’s instruction. After 4 hrs, the cells were lysed with 500 μl of lysis buffer (Promega). Forty μl of the supernatant were used for the luciferase assays using the Dual-Luciferase® Reporter Assay System (Promega) and analyzed in a Luminoskan luminometer.
Western blot analysis
Cells were harvested in cell lysis buffer (Promega) with protease (Roche) and phosphatase inhibitors (Pierce). The cell lysate was analyzed by Western blot by separating the proteins on a 12% SDS-polyacrylamide gel, with subsequent transfer onto nitrocellulose. The membranes were blocked and probed with an antibody directed against Aurora A (35C1, Calbiochem), eEF2K (Cell Signaling), eIF4E (BD Transduction), Gapdh (FL-335) (Santa Cruz Biotechnology), 4E-BP1 (53H11, Cell Signaling), Phospho-4E-BP1 (Thr37/46, Cell Signaling), or eIF-4G (C45A4, Cell Signaling) in 5% nonfat dried milk in a solution of PBS containing 0.1% Tween 20. The blots were then incubated with HRP-conjugated secondary antibodies (Promega). Immunoreactive bands were detected using Amersham® ECL plus (GE Healthcare) and quantified using ImageQuant version 5.2 (Molecular Dynamics) or Image J software (NIH).
Polysome analysis
Cells were treated with cycloheximide (50 ng/mL) for thirty minutes, then harvested on ice in PBS containing 50 ng/mL of cycloheximide and finally lysed with 400 μL of 100 mM KCL, 50 mM Tris-Cl, 1.5 mM MgCl2, 1mM DTT, 1.5% NP-40, protease inhibitors (Roche), 100 μg/mL cycloheximide and 100 U RNasin® plus RNase inhibitor (Promega). Then 300 μL of lysate was loaded on a 20–60% sucrose gradient created using a BIOCOMP Gradient Station and centrifuged at 39,000 rpm for 2 hrs at 4 °C using a SW40Ti rotor (Beckman Coulter). The gradient was fractionated and RNA was extracted from each fraction using TRIzol® Reagent (Sigma) followed by PureLink™ RNA Mini Kit (Invitrogen) and analyzed by qRT-PCR.
RNA extraction/qRT-PCR
Total RNA was extracted using TRIzol® Reagent (Sigma) followed by PureLink™ RNA Mini Kit (Invitrogen). cDNA libraries were synthesized using iScript™ cDNA Synthesis Kit (Bio-Rad), including a (−)RT control. The primer pairs used for the qRT-PCR were as follows: 5′-TCTTCACAGGAGGCAAATCCA-3′ (forward) and 5′-AATAAGTTACACACTCACTCAGGTACTA-3′ (reverse) for Aurora A mRNA, 5′-ACAGTCAGCCGCATCTTCTT-3′ (forward) and 5′-GTTAAAAGCAGCCCTGGTGA-3′ (reverse) for Gapdh mRNA and 5′-AAAGCTCCCAATCATCCAAA-3′ (forward) and 5′-GAGATGTGACGAACGTGT-3′ (reverse) for Photinus luciferase mRNA. qRT-PCR was performed using a Roche Lightcycler® 480 with either LightCycler® 480 SYBR Green I Master (Roche) or SsoAdvanced™ SYBR® Green Supermix (Bio-Rad) per manufacturer’s instructions.
RESULTS
Enhanced protein synthesis contributes to over-expression of Aurora A kinase
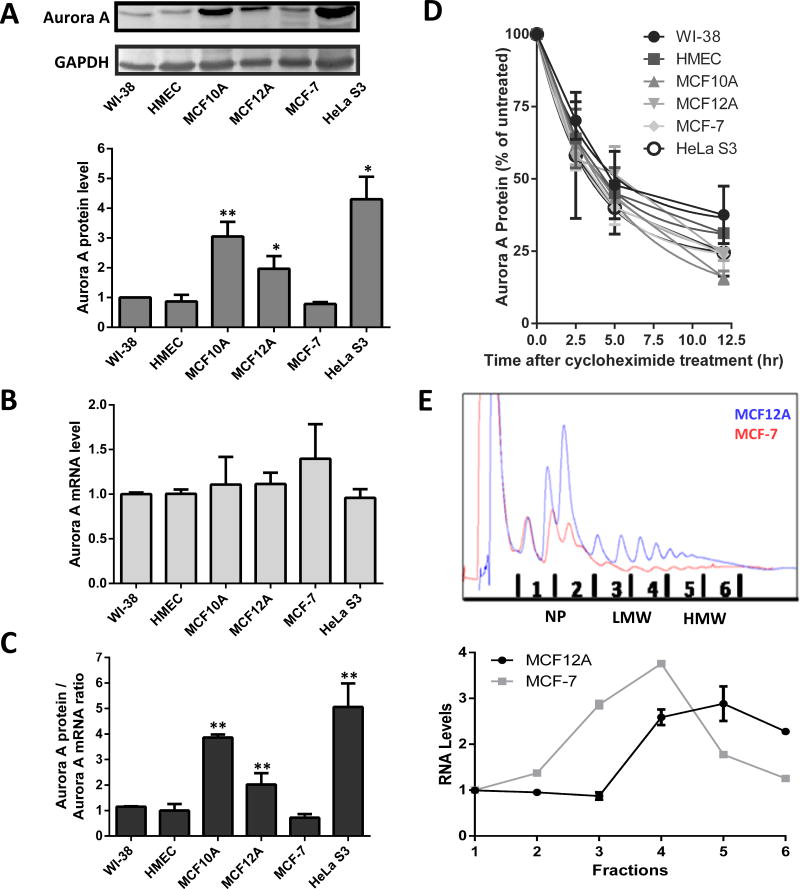
Increased Aurora A kinase expression is proposed to contribute to cellular immortalization and to the epithelial-mesenchymal transition (36,37). To identify mechanisms contributing to this enhancement, Aurora A protein expression was examined in a variety of cells, focusing on breast epithelial cell lines. Aurora A protein levels were quantified in two finite lines, normal human lung fibroblasts (WI-38) and human mammary epithelial cells (HMEC). Aurora A protein expression in these cells was chosen to represent basal protein levels and compared to the following immortalized cell lines: non-tumorigenic breast epithelial cell lines (MCF10A and MCF12A), and tumorigenic epithelial cell lines (MCF-7 and HeLa S3) from breast and cervix, respectively. Lysates were analyzed via Western blotting for Aurora A and Gapdh (as a loading control). Expression of Aurora A kinase in the control cell lines, WI-38 and HMEC, were similar and comparable to that observed in MCF-7 cells (Figure 1A). However, Aurora A kinase expression was 2 to 4.2 fold higher in MCF10A, MCF12A and HeLa S3 cells (Figure 1A). Three out of the 4 immortalized cell lines demonstrated Aurora A protein levels similar to those observed in breast and cervical cancer (36).
Figure 1.
Enhanced translation contributes to over-expression of Aurora A protein in a subset of immortalized cell lines. (A) Endogenous Aurora A protein levels were analyzed via Western blotting and normalized to Gapdh. The Aurora A/Gapdh ratio from each cell line was compared to the ratio from WI-38 which was set to one. n=6 ± standard deviations (SD) (B) Total RNA was isolated and analyzed via qRT-PCR with Gapdh as a reference target. Results were normalized to the Aurora A mRNA level in WI-38 cells. n=3 ± SD (C) The ratio of the normalized Aurora A protein levels to normalized Aurora A mRNA levels are shown. (D) Quantitation from Western blotting of Aurora A protein expression levels in cells treated for 0 to 12.5 hours with cycloheximide. Expression level of Aurora A protein in the untreated cells (0 hr) was normalized to 100. n=3 ± SD (E) UV detection readout of sucrose gradient fractionation of MCF12A and MCF-7 cells (top). Quantitation of Aurora A mRNA levels associated with the different gradient fractions. Levels were measured using qRT-PCR after isolation of total RNA from each fraction (bottom). * = p<0.05, ** = p<0.005, *** = p<0.0001, student’s t test.
The Aurora A gene is transcribed and the mRNA is translated during late S phase and peaks during G2/M phase of the cell cycle (4,5). Therefore, it is possible that differences in the number of cells in these phases from an asynchronous population could contribute to the observed differences in Aurora A protein levels. However, FACS analysis of the cell lines did not show a correlation with the percentage of cells in G2/M and Aurora A protein levels (Supplemental Figure 2). Therefore alterations in transcription, translation and/or protein stability are potentially contributing to increased Aurora A kinase expression in MCF10A, MCF12A and HeLa S3 cells.
To quantify Aurora A mRNA levels, cDNA libraries were constructed from total RNA isolated from the individual cell lines. Aurora A transcript levels from each cell line were measured by qRT-PCR and showed that mRNA levels were similar between each cell line except for a modest increase in MCF-7 cells (Figure 1B). The ratio of Aurora A protein to mRNA within each cell line was significantly elevated (ranging from approximately 2–5 fold higher) in MCF10A, MCF12A and HeLa S3 cells compared to WI-38 and HMEC cells, and even higher when compared MCF-7 cells (Figure 1C). These results indicate that increased Aurora A protein expression in the MCF10A, MCF12A and HeLa S3 cells is not due to enhanced transcription of the Aurora A gene or Aurora A mRNA stability, but the result of increased protein synthesis and/or protein stability.
To identify if alterations in protein stability contributed to the differential Aurora A expression, the half-life of the Aurora A protein was determined. Cells were harvested at 2 – 12 hours (hrs) after being treated with cycloheximide. The Aurora A protein levels were then analyzed via Western blotting (Supplemental Figure 3) with Gapdh as a loading control since it has a half-life of 90 to 120 hr (38). The half-life of the Aurora A protein was similar between the cell lines, ranging between 2.2 to 2.9 hr (Figure 1D). This range is consistent with a previous study that found the Aurora A protein half-life to be approximately 2.5 hrs (5). In addition, this result indicates that differences in protein stability do not contribute to the differential expression of Aurora A protein.
By order of elimination, the previous results implicated protein synthesis as one of the remaining mechanisms contributing to the variable Aurora A protein levels. To confirm this hypothesis, a polysome gradient analysis was performed. Aurora A mRNA levels were quantified in nonpolysomal, low molecular weight (LMW) polysome and high molecular weight (HMW) polysome fractions between the low Aurora A protein expressing MCF-7 cells and high Aurora A protein expressing MCF12A cells. Association with HMW fractions suggests increased translation initiation and/or reinitiation as the result of more efficient loading of the ribosomes onto the mRNA (39). Aurora A mRNA was most concentrated in the HMW fractions from MCF12A cells. In contrast, the majority of Aurora A mRNA levels from MCF-7 cells resided with the LMW fractions (Figure 1E). These results indicated translational up-regulation as a contributing mechanism to enhanced protein expression in MCF12A cells compared to MCF-7 cells.
Cap-dependent translation initiation is increased in the immortalized cell lines
The major mechanism by which translation is regulated is at the step of initiation. And the primary mode by which translation is initiated is through recognition of the m7G cap structure (40,41). Misregulating cap-dependent translation can lead to elevated levels of protein synthesis and tumorigenesis. For example, increased expression of the rate-limiting canonical factor eIF-4E (which binds the cap structure) is found in many tumors (42–44). Furthermore, ectopically induced over-expression of eIF-4E has transforming capabilities leading to malignant phenotypes (45–47). To determine if elevated eIF-4E levels were contributing to the over-expression of Aurora A, Western blots of lysates were analyzed. eIF-4E levels were similar in all six cell lines (Supplemental Figure 4A). This observation suggests that a global increase in cap-dependent translation may not be occurring. On the other hand, the eIF-4E that is present may be differentially restricted in its ability to bind the cap structure.
The 4E binding protein 1 (4E-BP1), a negative regulator of eIF-4E, is often decreased in cancer cells (48). Analysis of 4E-BP1 protein levels via Western blotting showed there was no difference between 4E-BP1 levels in WI-38, MCF12A or MCF-7 cells (Supplemental Figure 4B). However, there was actually a 1.5 to 2.2 fold increase in the expression of 4E-BP1 in HMEC, MCF10A, and HeLa S3 cells. The phosphorylation state of 4E-BP1 is integral to its activity with the hypophosphorylated 4E-BP1 able to bind 4E and inhibit cap-dependent translation. Interestingly, there was a 1.5 to 3 fold increase of hypophosphorylated 4E-BP1 in all the cell lines compared to WI-38 cells (Supplemental Figure 4B right). Taken together, these results do not support the hypothesis that cap structure accessibility is altered in the different cell lines.
The major scaffolding protein eIF-4G is the key structural component in the pre-initiation complex. Its expression level is proposed to contribute to the rate of translation (49). However, eIF-4G levels did not correlate with Aurora A protein levels as demonstrated by the increased expression of eIF-4G in MCF-7 cells compared to WI-38 and HMEC cells (Supplemental Figure 4C). Additionally eIF-4G protein expression was low in MCF-12A cells.
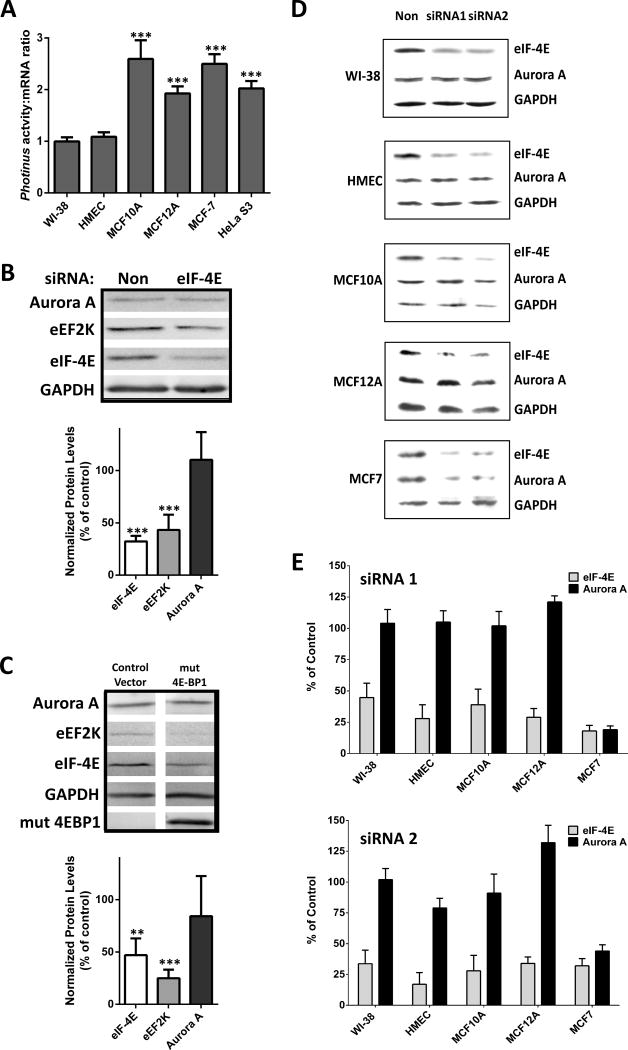
The expression patterns of the critical cap-dependent regulatory proteins eIF-4E, 4EBP, and eIF-4G did not yield any candidates that may be contributing to the over-expression of Aurora A kinase. To definitively compare cap-dependent translation initiation between these cell lines we utilized a reporter assay. In vitro transcribed mRNA from monocistronic DNA containing the Photinus luciferase open reading frame (ORF), the β-globin 5′ leader and the SV40 poly-adenylation (A) site was created. The β-globin 5′ leader was chosen because it is short (53 nt), unstructured, without an upstream ORF (uORF), and the β-globin mRNA is exclusively translated in a cap-dependent manner (41). The transcripts were capped with a m7G cap, poly (A) tailed, and transfected into each cell line. After 4 hrs the cells were harvested and assayed for Photinus luciferase. The luciferase activity from each cell line was compared to transcript levels quantitated using qRT-PCR to normalize for transfection efficiency as well as mRNA stability. Translation of the reporter was enhanced by 1.9 to 2.6 fold in all of the immortalized cell lines compared to the normal cells (Figure 2A). Interestingly, MCF-7 cells, which do not over-express the Aurora A protein, elicited the second largest increase of cap-dependent translation compared to WI-38 and HMEC cells (Figure 2A). This result indicates that other mechanisms aside from eIF-4E, 4E-BP, and eIF-4G expression are contributing to enhanced cap-dependent translation. However, increased cap-dependent translation did not correlate with increased expression of Aurora A protein, thereby suggesting an alternate mechanism may be regulating translation of the Aurora A mRNA.
Figure 2.
Inhibiting cap-dependent translation initiation differentially affects Aurora A protein expression. (A) Translation of a Photinus luciferase reporter mRNA containing the β-globin 5′ leader was transfected into the six cell lines. Luciferase activity was measured and the mRNA quantitated by qRT-PCR after 7 hrs. Shown is the ratio of luciferase activity to luciferase RNA. n=3 in triplicate ± SD (B) Western blots of lysates from HeLa cells transfected with nonsense siRNA or siRNA targeting eIF-4E for 48 hr. n=3, ± SD (C) Western blots of lysates from HeLa cells transfected with a control vector or a plasmid encoding for mutant hypophosphorylated 4E-BP1 for 48 hr. n=3 ± SD (D) Western blots of lysates from WI-38, HMEC, MCF10A, MCF12A and MCF-7 cells transfected with nonsense siRNA, siRNA 1 targeting eIF-4E or siRNA 2 targeting eIF-4E for 48 hr. (E) Quantification of eIF-4E knockdowns in WI-38, HMEC, MCF10A, MCF12A and MCF-7 cells. n=3 ± SD, ** = p<0.005, *** = p<0.0001, student’s t test.
Aurora A protein expression level is unaffected by inhibiting cap-dependent translation initiation
To further determine the role of cap-dependent translation initiation in the synthesis of the Aurora A protein, cap-dependent translation was inhibited using two different approaches. Initially eIF-4E expression was knocked down. HeLa cells were transfected with siRNA (Dharmacon) targeting eIF-4E mRNA or nonsense siRNA for 48 hr. Quantification of Western blots showed that eIF-4E protein expression was reduced by nearly 70% (Figure 2B). Eukaryotic elongation factor 2 kinase (eEF2K) expression in the eIF-4E siRNA treated cells decreased by 57%, similar to previous observations (50). In contrast, the level of Aurora A protein was equivalent in the eIF-4E and nonsense siRNA conditions (Figure 2B).
As a second approach to modulate cap-dependent translation, HeLa cells were transfected with a DNA plasmid encoding a hypophosphorylated mutant of 4E-BP1 in which the main five phosphorylation sites are mutated rendering the protein non-phosphorylated (generously provided by Dr. Davide Ruggero, University of California, San Francisco) or a control vector (Paltag). The constitutively expressed hypophosphorylated 4E-BP1 should sequester eIF-4E, thereby disrupting eIF-4F formation and inhibit cap-dependent translation (51). 48 hrs after transfection, cells were harvested and protein levels were analyzed by Western blotting. eEF2K expression decreased by 75% in the presence of hypophosphorylated 4E-BP1 (Figure 2C). eIF-4E expression decreased by 53%, similar to previous results (50), indicating that the eIF-4E mRNA is translated via a cap-dependent mechanism and a decrease in cap-dependent translation has occurred. On the other hand, the Aurora A protein level decreased by only 16% (Figure 2C). Taken together, these results show that synthesis of Aurora A kinase protein is relatively unaffected when cap-dependent translation is inhibited in HeLa cells indicating that an alternative mechanism may be contributing to the translation initiation of the Aurora A mRNA.
To determine if cap-dependent translation regulates expression of the Aurora A protein in other cells, two siRNAs targeting eIF-4E were individually transfected into WI-38, HMEC, MCF10A, MCF12A and MCF-7 cells. eIF-4E was significantly knocked down by each siRNA in all cell lines (Figure 2D–E). Aurora A protein expression was significantly decreased in MCF-7 cells, indicating cap-dependent initiation is the primary mechanism for translation of Aurora A mRNA in these cells. However, Aurora A protein levels were unaffected when eIF-4E levels were reduced in WI-38, HMEC, MCF10A and MCF12A cells (Figure 2D–E). Taken together, these results suggest an alternate mechanism may be utilized to initiate translation of the Aurora A mRNA.
The Aurora A 5′ leader contains an IRES
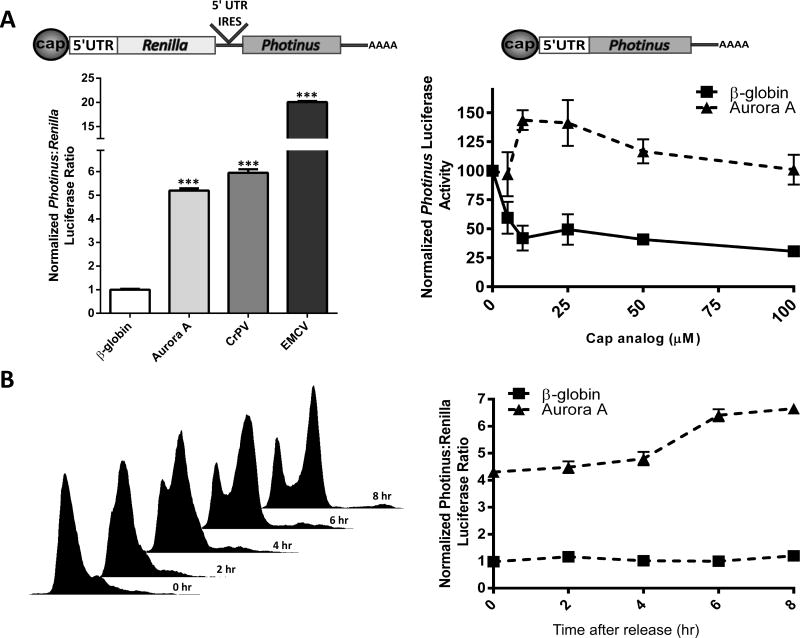
Cap-dependent translation did not appear to be a significant mechanism for the synthesis of Aurora A protein in the HMEC cells and in the cell lines in which Aurora A kinase was over-expressed. This result indicated it may be translated in a cap-independent manner. To determine if the Aurora A 5′ leader has an IRES, it was inserted into the intercistronic region of a dicistronic luciferase construct coding for Renilla and Photinus luciferase in the first and second cistrons, respectively (29). The β-globin 5′ leader, which does not contain an IRES, was used as a negative control. Two viral IRESes, the encephalomyocarditis virus (EMCV) IRES and the IRES in the intergenic region of the cricket paralysis virus (CrPV) were chosen as positive controls (52,53) The constructs were in vitro transcribed to eliminate the possibility of cryptic promoter activity or alternative splicing (54,55). The resulting dicistronic transcripts were transfected into HeLa cells. After 7 hrs the cells were harvested and assayed for Renilla and Photinus luciferase. The Photinus:Renilla (P:R) luciferase ratio obtained from the β-globin dicistronic construct was normalized to one. Consequently, a ratio above one would indicate IRES activity. The P:R ratio obtained from the dicistronic luciferase mRNA containing the Aurora A 5′ leader was 5 fold higher than that obtained from the dicistronic mRNA containing the β-globin 5′ leader (Figure 3A left). This ratio was similar to the one obtained from the mRNA containing the CrPV IRES, but less than the 20 fold increase in the P:R ratio from the mRNA containing the EMCV IRES. These results indicated that the Aurora A 5′ leader contains an IRES which exhibits activity similar to the CrPV IRES in HeLa cells.
Figure 3.
The 5′ leader of the Aurora A mRNA contains an IRES. (A) Dicistronic luciferase mRNA (left) containing the β-globin, Aurora A, CrPV, and EMCV 5′ leaders or IRES elements were transfected individually into HeLa cells. Luciferase activity is shown as the ratio of Photinus luciferase to Renilla luciferase (P:R) and is normalized to that obtained from the dicistronic mRNA containing the β-globin 5′ leader. n=3 in triplicate ± SD Monocistronic Photinus luciferase mRNA (right) containing the β-globin or Aurora A 5′ leader were translated in rabbit reticulocyte lysate in the presence of increasing concentrations of cap analog. The initial level of Photinus luciferase activity from each monocistronic mRNA was normalized to 100. n=3 ± SD (B) Cell cycle progression following the release of a double thymidine block in HeLa cells was confirmed by flow cytometry analysis (left). HeLa cells transfected with dicistronic luciferase mRNA containing the β-globin and Aurora A 5′ leaders were synchronized in G1/S with a double thymidine block and released. Luciferase activity is shown as the ratio of Photinus luciferase to Renilla luciferase (P:R) and is normalized to that obtained from the dicistronic mRNA containing the β-globin 5′ leader at the 0 time point (right). n=3 in triplicate ± SD, *** = p<0.0001, student’s t test.
To further validate the presence of an IRES in the Aurora A 5′ leader, in vitro transcribed monocistronic mRNA containing the Photinus luciferase ORF and the β-globin or Aurora A 5′ leader was translated in rabbit reticulocyte lysate. Cap-dependent translation was inhibited by adding increasing amounts of cap analog to compete with the m7G cap. Luciferase activity measured from the β-globin construct decreased by 70% in the presence of 100μM of cap analog while luciferase activity from the Aurora A construct was relatively unchanged (Figure 3A right). This result demonstrated that the Aurora A 5′ leader initiates translation independently of the cap structure and supports the hypothesis that it contains an IRES.
Expression levels of both Aurora A mRNA and Aurora A protein peak during the G2/M phase of the cell cycle (4,5). This temporal expression coincides with reports indicating that IRES activity is regulated through the cell cycle and peaks at G2/M (56,57). To determine if the Aurora A IRES is regulated by the cell cycle, HeLa cells were synchronized by a double thymidine block. During the last thymidine block the cells were transfected with in vitro transcribed dicistronic mRNA containing either the β-globin or Aurora A 5′ leader in the intercistronic region. Cells were harvested between 0 to 8 hrs following release from the double thymidine block into normal growth medium with the total transfection time being 11 hrs for all cells. FACs analysis showed that the majority of cells were synchronized in G1/S and proceeded through the cell cycle, entering G2/M at approximately 6 to 8 hrs after release (Figure 3B left). The P:R ratio obtained from the β-globin construct at the time of thymidine release was set to one and did not change during the phases of the cell cycle. On the other hand, immediately after thymidine release the Aurora A P:R ratio was 4.2 fold higher than that obtained with β-globin. The maximum P:R ratio, a 6.4 fold increase occurred 8 hrs later (Figure 3B right). This result indicates that the Aurora A IRES is active throughout the cell cycle but peaks during the same portion of the cell cycle in which translation of the endogenous Aurora A mRNA does – G2/M.
Aurora A IRES activity is increased in the cell lines that over-express Aurora A protein
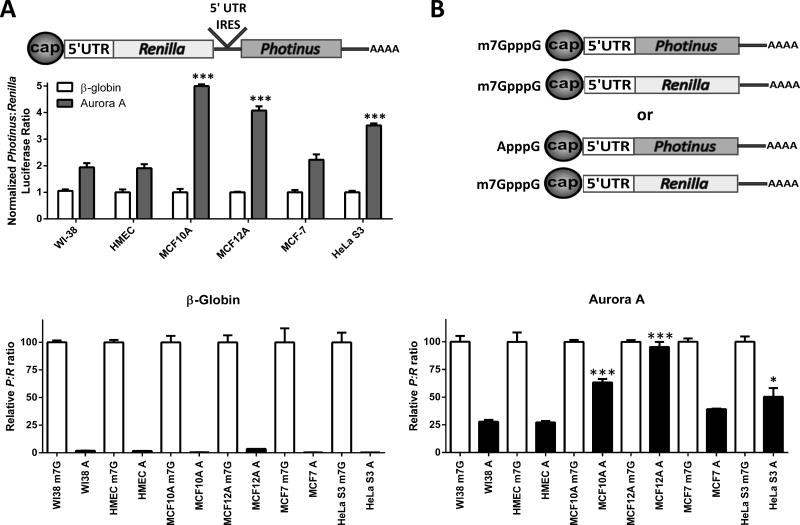
To determine if Aurora A IRES activity parallels Aurora A protein expression, dicistronic mRNA containing the Aurora A or β-globin 5′leaders was transfected into all six cell lines. After 7 hrs the cells were harvested and assayed for Renilla and Photinus luciferase. The P:R ratio for the mRNA containing the Aurora A 5′ leader was at least two-fold higher than the control in the six cell lines indicating the Aurora A IRES is operative in all cells (Figure 4A). Moreover, the Aurora A P:R ratio was highest in the cell lines that also over-expressed the Aurora A protein (Figure 4A). These results show a positive correlation between Aurora A protein expression and Aurora A IRES activity. This correlation supports the hypothesis that IRES-dependent translation is contributing to the over-expression of the Aurora A protein observed in the subset of cell lines.
Figure 4.
Aurora A IRES activity is increased in the subset of immortalized cell lines that exhibit enhanced Aurora A protein expression. A) Dicistronic mRNA containing the β-globin and Aurora 5′ leaders were transfected into each cell line. The P:R ratio obtained from the β-globin construct from each cell line was set to one. The P:R ratio from the Aurora A construct was normalized to β-globin. n=3 in triplicate ± SD (B) Schematic representation of cotransfections of an A or m7G capped monocistronic Photinus luciferase mRNA containing the Aurora A or β-globin 5′ leaders with an m7G capped Renilla luciferase mRNA. C) Normalized luciferase activity obtained from the A capped Photinus luciferase constructs are represented as a percentage of the normalized luciferase activity obtained from the m7G capped Photinus luciferase construct for the β-globin 5′ leader (left) or the Aurora A 5′ leader (right). n=3 in triplicate ± SD, * = p<0.05, *** = p<0.0001, student’s t test.
All eukaryotic transcripts contain a methyl-7-guanosine cap. Accordingly, translation of the Aurora A mRNA may be initiated through the cap structure and/or the IRES. To quantitate the contribution of the Aurora A IRES in a monocistronic mRNA, the β-globin or Aurora A 5′ leader were placed upstream of the Photinus luciferase ORF and in vitro transcribed. The transcripts were capped with an m7G or ApppG cap (A cap) and poly (A) tailed (Figure 4B). The A cap is not recognized by eIF-4E preventing cap-dependent translation initiation of the mRNA. In addition, the poly (A) specific 3′ exonuclease PARN does not recognize the A cap, thereby inhibiting deadenylation and preventing both 5′ and 3′ exonucleolytic mRNA decay (58,59). The mRNA was co-transfected into each cell line with a m7G capped and poly (A) tailed mRNA containing the humanized Renilla luciferase ORF to normalize for transfection efficiency. For each cell line, the P:R ratio obtained from the m7G cap Photinus constructs was set to 100. The P:R ratio obtained from the A cap Photinus luciferase mRNA was compared to the P:R ratio obtained from the corresponding m7G cap Photinus luciferase constructs. The A cap reporter mRNA containing the β-globin leader was poorly translated (5 to 16% of the m7G cap P:R ratio) (Figure 4C left). This result demonstrated that the translation of the mRNA containing the β-globin 5′ leader was dependent on the presence of the m7G cap structure. In the WI-38 and HMEC cells, translation of the A cap mRNA containing the Aurora A 5′ leader was higher than the mRNA with the β-globin leader. This result indicates that the Aurora A 5′ leader was initiating cap-independent translation, although it was only approximately 25% of that obtained by a m7G cap mRNA containing the Aurora A 5′ leader (Figure 4C right). On the other hand, the A cap P:R ratio from the Aurora A construct was considerably higher in the cell lines that over-express the Aurora A protein (MCF10A, MCF12A, and HeLa S3). Remarkably, in the MCF12A cells Photinus luciferase activity from the A cap mRNA was 95% of that obtained from the m7G cap Aurora A mRNA (Figure 4C). These results suggest cap-dependent translation is most likely a contributing mechanism for Aurora A protein synthesis in the low expressing lines, but IRES-dependent translation is the predominant mechanism in the Aurora A over-expressing lines.
To determine if IRES activity in general was upregulated in the Aurora A over-expressing cell lines, the experiment was repeated using IRESes from the PITSLRE kinase, EMCV, and FMR1 mRNAs. Whereas all 3 IRESes are utilized in proliferating cells, PITSLRE and EMCV IRES activity is upregulated during G2/M phase of the cell cycle while the FMR1 IRES is predominantly employed in post-mitotic neurons (50,60). Internal initiation mediated by the FMR1 IRES varied between the cell lines but the activity did not correlate with that observed from the Aurora A IRES (Supplemental Figure 5A). Compared to results in HMEC cells, FMR1 IRES activity was similar in MCF12A and MCF-7 cell lines, significantly elevated in MCF10A, slightly decreased in WI-38 cells, and significantly decreased in HeLa S3 cells. Interestingly, activity from the EMCV and PITSLRE IRESes also did not correlate with that observed from the Aurora A IRES (Supplemental Figure 5B and C, respectively). EMCV IRES activity was significantly lower in MCF10A and MCF-7 cells compared to HMEC cells. There was no difference in activity between WI-38, HMEC, MCF12A and HeLa S3 cells. On the other hand, activity from the PITSLRE IRES was significantly elevated in MCF12A and HeLa S3 cells yet significantly decreased in MCF-7 cells compared to the normal cells, similar to Aurora A IRES activity. However, unlike the Aurora A IRES, the PITSLRE IRES was less active in MCF10A cells. In addition, PITSLRE IRES activity was higher in the WI-38 and HMEC cells compared to Aurora A IRES activity. These results indicate that not all IRESes are upregulated in the Aurora A over-expressing cell lines. Furthermore, they suggest possible variations in the regulation of IRESes that are upregulated under similar circumstances, such as the G2/M phase of the cell cycle.
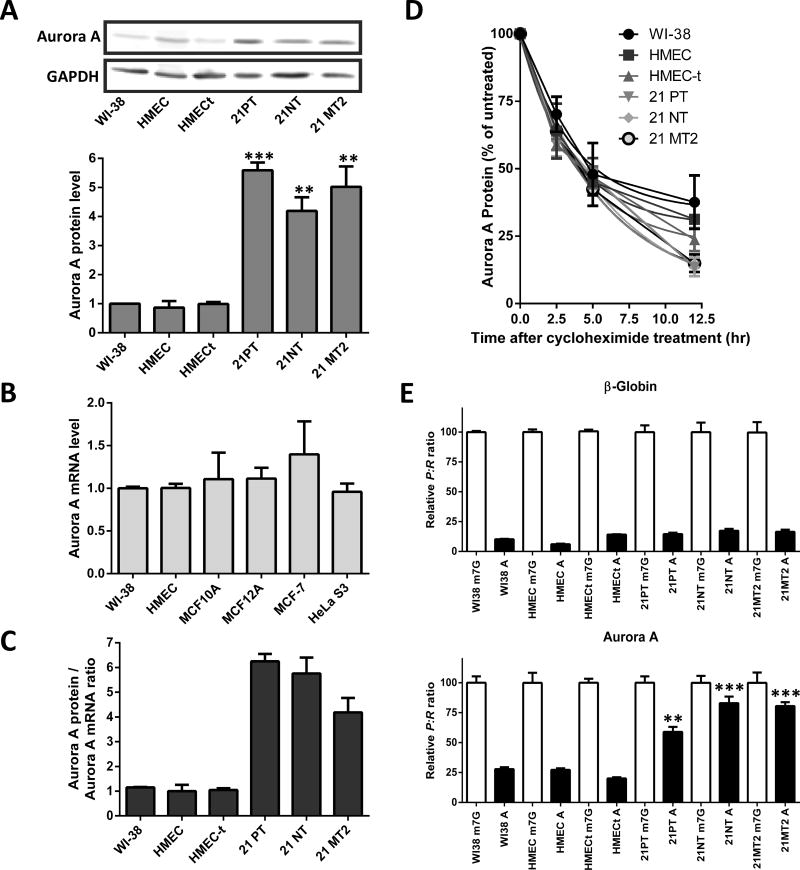
To determine if the correlation between Aurora A IRES activity and protein expression extends to other cell lines, we performed the same assay with three lines from the 21T cell line series (21PT, 21NT, and 21MT2). These cell lines were derived from a single patient diagnosed with infiltrating and intraductal mammary carcinoma and are often used as an in vitro model for cancer progression. The non-tumorigenic 21PT and the tumorigenic 21NT cell lines were derived from a primary tumor. The 21MT2 line was derived from a metastatic tumor (61–63). All three cell lines over-express Aurora A protein (Figure 5A), yet mRNA levels and protein half-life (2.8–3.1 hrs) were equivalent to the normal cell lines (Figures 5B and 5D). The elevated Aurora A protein/mRNA ratio indicated an increase in Aurora A translation (Figure 5C). And the A/m7G cap monocistronic transfection experiment demonstrated a high level of Aurora A IRES usage. The percentage of A cap mediated translation to that obtained from the m7G cap with the Aurora A 5′ leader ranged from 60% (21PT) to 83–85% (21MT2 and 21NT respectively) (Figure 5E bottom). These results are similar to what was observed with the other Aurora-A over-expressing cell lines.
Figure 5.
Additional breast epithelial cell lines demonstrate a correlation between Aurora A IRES activity and Aurora A protein expression. (A) Endogenous Aurora A protein levels were analyzed via Western blotting and normalized to Gapdh. The Aurora A/Gapdh protein ratio from each cell line was compared to the ratio from WI-38 which was set to one. n=3 ± SD (B) Total RNA was isolated from the four cell lines and analyzed via qRT-PCR with Gapdh as a reference target. Results were normalized to the Aurora A mRNA level in WI-38 cells. n=3 ± SD (C) The ratio of the normalized Aurora A protein levels to normalized Aurora A mRNA levels are shown. (D) Quantitation from Western blotting of Aurora A protein expression levels from the four cell lines treated for 0 to 12.5 hours with cycloheximide. Expression level of Aurora A protein in the untreated cells (0 hr) was normalized to 100. n=3 ± SD (E) Normalized luciferase activity from the A capped Photinus luciferase constructs are represented as a percentage of the normalized luciferase activity obtained from the m7G capped Photinus luciferase construct for the β-globin 5′ leader (top) or the Aurora A 5′leader (bottom). n=3 in triplicate ± SD, ** = p<0.005, *** = p<0.0001, student’s t test.
To resolve whether increased proliferative activity contributed to alterations in Aurora A IRES activity, this assay was performed in a HMEC cell line that ectopically expresses telomerase (HMEC-t); a cellular process that prevents replicative cellular aging. Over-expressing telomerase did not alter Aurora A protein levels, mRNA levels, or protein stability (Figures 5A–5D). In addition, the A cap P:R ratio exhibited by these cells was similar to that obtained from WI-38 and HMEC cells (Figure 5E).
In summary, these results demonstrate that IRES-dependent translation initiation mediated by the Aurora A 5′leader is utilized to a greater extent in cells over-expressing the Aurora A protein. Simply extending proliferative activity does not alter Aurora A IRES activity or protein levels. These results support the hypothesis that enhanced IRES-dependent translation of the Aurora A mRNA contributes to the increase of Aurora A kinase expression, which predisposes the cell to immortalization and tumorigenesis.
DISCUSSION
Over-expression of the Aurora A kinase is observed in a broad range of malignancies including breast, brain and pancreatic tumors (8–10,64–66). Misregulation of its expression is proposed to be both an early event in the immortalization of the cell as well as a contributor to the epithelial–mesenchymal transition (EMT) (37,67). In the present report, the goal was to identify mechanism(s) contributing to Aurora A protein over-expression. To this end we chose six cell lines, which included primary, immortalized, and tumorigenic cell types. They were categorized as either high or low Aurora A protein expressing cells. Transcription/mRNA stability, protein stability, nor cap-dependent translation could account for enhanced levels of Aurora A protein in the high expressing lines. On the other hand, we identified an IRES in the Aurora A 5′ leader and IRES activity strongly correlated with Aurora A protein expression.
Utilizing RNA dicistronic luciferase constructs, Aurora A IRES activity was shown to correlate with Aurora A protein expression. However, this assay does not measure the contribution of the IRES in an m7G capped monocistronic mRNA, the state in which the Aurora A mRNA is present. It has been suggested that eukaryotic IRESes initiate translation at a considerably reduced rate compared to m7G cap-dependent initiation (21,68). Thus, even if IRES activity is enhanced in a subset of cell lines the overall increase may not be physiologically significant. To address this issue, we created monocistronic mRNA containing the Aurora A IRES with an A or m7G cap. These RNAs would permit comparison between translation initiated in a cap-independent manner to translation mediated by an m7G cap and an IRES (58). The resulting A cap/m7G cap ratio we termed ‘IRES usage’. In WI-38 and HMEC cells, A-capped translation of the RNA containing the Aurora 5′ leader was less than 25% of that obtained from an m7G capped mRNA (Figure 4C). This result indicated that cap-dependent translation is likely the predominant mechanism. However, transfection into the other cell lines yielded dramatically different results. In MCF12A cells, translation of the mRNA was similar irrespective of the cap structure (Figure 4C). This is one of the first examples whereby a eukaryotic IRES initiates translation at a rate similar to an m7G capped transcript. We interpret this result as demonstrating that IRES-dependent translation is elevated and is the principal mechanism utilized to initiate translation of the m7G capped mRNAs. Alternatively, reduced cap-dependent translation without any concomitant alteration in IRES activity could yield a similar result. To differentiate between these two explanations, we examined cap-dependent translation of a monocistronic mRNA and found that it was relatively similar between the four immortalized lines and actually higher than the two primary cell lines (Figure 2A). Accordingly, these results indicate an apparent switch from low level cap-dependent translation of the Aurora A mRNA in normal, finite lifespan cells to increased IRES-dependent translation in immortalized cells prior to malignant transformation. Identifying the mechanism(s) that play a role in this switch could be crucial to understanding oncogenesis mediated by Aurora A kinase over-expression and possibly of other oncogenes which are translated in an IRES-dependent manner. For example, a subset of breast tumors exhibit over-expression of Her2 and EGF has been implicated in affecting Aurora A kinase expression (67).
Examination of additional cell lines reinforced the positive correlation between IRES usage and Aurora A protein expression. The 21T series exhibited both elevated Aurora A protein levels and IRES activity; while Aurora mRNA and protein half-life was similar to the finite cell lines (see Figure 5). This group of cells included immortalized, but nontumorigenic (21PT), tumorigenic (21NT), and finally metastatic (21MT2). These results are consistent with previous reports suggesting enhanced Aurora A expression contributes to immortalization and continues through the epithelial-mesenchymal transition (37,67).
Internal initiation of translation has been invoked as a mechanism involved in carcinogenesis (reviewed in (69)). Multiple mRNAs encoding proteins contributing to cell proliferation (FGF2, PDGF2), cell cycle (PITSLRE p58), cell death/survival (Bcl-X, Apaf1), and tumor development (p53, c-myc, c-jun) contain IRESes (28,29,32,70–73). Misregulating IRES-dependent translation contributes to cancer progression through multiple means. For example, in the disease X-Linked Dyskeratosis Congenita, mutations in the dyskerin gene reduce pseudouridylation of rRNA, which inhibits IRES-dependent translation of tumor suppressors (p53, p27) and apoptotic factors (Bcl-X, XIAP) predisposing individuals to cancer (74–76). Alternatively, the low oxygen environment in the center of solid tumors increases IRES-dependent translation of the VEGF mRNA, which in turn promotes angiogenesis and tumor growth (77). In the present study, we found that the Aurora A IRES is regulated through the cell cycle and by cell type. As a first step towards identifying the mechanism, other IRESes that are upregulated in G2/M were examined. However, there was no correlation of IRES usage between the PITSLRE, EMCV and the Aurora A IRES. It would be of interest to determine if there is a subset of IRESes, perhaps those encoding other oncogenes, which exhibit IRES usage patterns similar to Aurora A.
The mechanism regulating the Aurora A IRES is unknown. Identifying cis-elements that control IRES activity would be informative. Viral IRESes generally rely on extensive secondary structure, a feature that is often conserved phylogenetically, although the sequences can vary (78–80). The presence of uORFs are observed as well (23,81). Alternatively, eukaryotic IRESes are quite variable; sequence and/or secondary structure does not imply the presence of an IRES. Previous studies have reported eukaryotic IRES motifs as short as 9 – 50 nt (82,83), but they can extend to over 1000 nt (84).
Viral IRESes exhibit a variable requirement for trans-factors. Some IRESes can recruit the ribosome directly while others utilize both canonical factors (including eIF-4E in certain situations) and IRES-transacting factors (ITAFs) such as polypyrimidine-tract binding protein or La protein (52,85–87). The rate-limiting step for eukaryotic IRESes is proposed to be the expression level of ITAFs (reviewed in (88)). These are RNA binding proteins that aid in the recruitment of the translational machinery and/or alter the RNA secondary structure, which in turn promotes binding of the translation complex. ITAF mis-expression can alter translation of cancer-related mRNAs. For example, murine double minute (MDM2) is an oncoprotein that binds the IRES in the mRNA encoding the X-linked inhibitor of apoptosis protein (XIAP). Increased XIAP expression in the MDM2 over-expressing cells leads to a resistance to radiation-induced apoptosis (89). Alternatively, a loss of two other ITAFs, TCP80 and RHA diminishes p53 IRES activity and protein expression; promoting cell survival in response to DNA damage (90). Presumably, there are ITAFs whose increased expression is responsible for the enhanced Aurora A IRES activity. Since there is no in vivo assay to quantify Aurora A IRES activity in normal/immortalized cells, the level of these regulatory proteins would be a useful biomarker and provide a novel drug target to modulate Aurora A IRES activity in cancer cells. Two ITAFs known to be expressed in G2/M are polypyrimidine-tract binding protein (PTB) and upstream of n-ras (Unr). They both bind the PISTRLE p58 IRES and regulate the G2/M specific expression of the PISTLRE p58 protein (57,91). They can also regulate IRESes during other phases of the cell cycle, such as the Apaf-1 IRES during G1 (35). As noted above there was no correlation between PITSLRE and Aurora A IRES usage. Furthermore, knocking down Unr did not affect Aurora A IRES activity (unpublished observations) and there does not appear to be any potential PTB binding sites in the Aurora A 5′ leader. Consequently, there are likely to be additional ITAFs that regulate the Aurora A and possibly other mitotic IRESes. Indeed, it would be of interest to determine whether the Aurora A IRES represents another subset of G2/M functionally related IRESes.
Current anti-cancer drugs target Aurora A kinase activity since its inhibition leads to formation of a monopolar spindle and cell death (92). However, it has been difficult to design reagents that exhibit specificity for the ATP pocket of the Aurora A kinase (93,94). Moreover, over-expressing a kinase dead mutant of Aurora A can still contribute to cellular immortalization (95).
Targeting the Aurora A mRNA could be an effective alternative approach to regulate expression of the kinase. HMEC (and WI-38 cells) yielded low Aurora A IRES activity, but surprisingly Aurora A protein expression was relatively unaffected when eIF-4E levels were reduced. This result indicates that endogenous Aurora A mRNA in normal cells (e.g. HMEC) can be translated in both a cap and IRES dependent manner. We postulate that the HMEC cells utilize both translation initiation mechanisms to ensure expression of the Aurora A kinase. Inhibiting cap-dependent translation in these cells is compensated by the normal up-regulation of IRES activity during G2/M. On the other hand, Aurora A expression was sensitive to the reduction of eIF-4E in MCF-7 cells. Aurora A IRES activity (and protein expression) remain low and cannot compensate for the reduction in cap-dependent translation. This result suggests that reducing cap-dependent translation would decrease endogenous Aurora A protein in tumors that cannot utilize the Aurora A IRES. Finally, IRES-dependent translation was increased in the Aurora A protein over-expressing cells and is the principal translation initiation mechanism. Consequently, designing reagents targeting the IRES would inhibit Aurora A protein expression primarily in cancer cells that over-express the kinase.
In summary, we have identified an IRES situated in the 5′ leader of the human Aurora A mRNA. In an examination of multiple cell lines IRES activity was the only mechanism that correlated with Aurora A protein expression. Moreover, this mechanism appears to be upregulated early during cancer development and remains elevated as cell transformation advances indicating that targeting this mechanism may be beneficial for multiple stages of cancer progression.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Jessica Tyler for helpful comments on the manuscript, Drs. Ford and DeGregori for cell lines and Dr. Ruggero for the 4E-BP1 mutant construct. We also thank Shihuang (Sam) Su and the other members of the Krushel lab for their crucial assistance. STR DNA fingerprinting was performed by the Cancer Center Support grant funded Characterized Cell Line core, NCI # CA16672.
FUNDING
This work was supported by the National Institutes of Health [AG02815G to LAK].
Footnotes
Disclosure: The authors state that they have no conflicts of interest in the present.
Supplementary Data are available at Molecular Cancer Research online: Supplementary Figures 1–5.
References
- 1.Adams RR, Carmena M, Earnshaw WC. Chromosomal passengers and the (aurora) ABCs of mitosis. Trends Cell Biol. 2001;11:49–54. doi: 10.1016/s0962-8924(00)01880-8. [DOI] [PubMed] [Google Scholar]
- 2.Nigg EA. Mitotic kinases as regulators of cell division and its checkpoints. Nat Rev Mol Cell Biol. 2001;2:21–32. doi: 10.1038/35048096. [DOI] [PubMed] [Google Scholar]
- 3.Fu J, Bian M, Jiang Q, Zhang C. Roles of Aurora kinases in mitosis and tumorigenesis. Mol Cancer Res. 2007;5:1–10. doi: 10.1158/1541-7786.MCR-06-0208. [DOI] [PubMed] [Google Scholar]
- 4.Tanaka M, Ueda A, Kanamori H, Ideguchi H, Yang J, Kitajima S, Ishigatsubo Y. Cell-cycle-dependent regulation of human aurora A transcription is mediated by periodic repression of E4TF1. J Biol Chem. 2002;277:10719–10726. doi: 10.1074/jbc.M108252200. [DOI] [PubMed] [Google Scholar]
- 5.Honda K, Mihara H, Kato Y, Yamaguchi A, Tanaka H, Yasuda H, Furukawa K, Urano T. Degradation of human Aurora2 protein kinase by the anaphase-promoting complex-ubiquitin-proteasome pathway. Oncogene. 2000;19:2812–2819. doi: 10.1038/sj.onc.1203609. [DOI] [PubMed] [Google Scholar]
- 6.Littlepage LE, Wu H, Andresson T, Deanehan JK, Amundadottir LT, Ruderman JV. Identification of phosphorylated residues that affect the activity of the mitotic kinase Aurora-A. Proc Natl Acad Sci U S A. 2002;99:15440–15445. doi: 10.1073/pnas.202606599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou H, Kuang J, Zhong L, Kuo WL, Gray JW, Sahin A, Brinkley BR, Sen S. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat Genet. 1998;20:189–193. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]
- 8.Bischoff JR, Anderson L, Zhu Y, Mossie K, Ng L, Souza B, Schryver B, Flanagan P, Clairvoyant F, Ginther C, et al. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. 1998;17:3052–3065. doi: 10.1093/emboj/17.11.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gritsko TM, Coppola D, Paciga JE, Yang L, Sun M, Shelley SA, Fiorica JV, Nicosia SV, Cheng JQ. Activation and overexpression of centrosome kinase BTAK/Aurora-A in human ovarian cancer. Clin Cancer Res. 2003;9:1420–1426. [PubMed] [Google Scholar]
- 10.Jeng YM, Peng SY, Lin CY, Hsu HC. Overexpression and amplification of Aurora-A in hepatocellular carcinoma. Clin Cancer Res. 2004;10:2065–2071. doi: 10.1158/1078-0432.ccr-1057-03. [DOI] [PubMed] [Google Scholar]
- 11.Sakakura C, Hagiwara A, Yasuoka R, Fujita Y, Nakanishi M, Masuda K, Shimomura K, Nakamura Y, Inazawa J, Abe T, et al. Tumour-amplified kinase BTAK is amplified and overexpressed in gastric cancers with possible involvement in aneuploid formation. Br J Cancer. 2001;84:824–831. doi: 10.1054/bjoc.2000.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goepfert TM, Adigun YE, Zhong L, Gay J, Medina D, Brinkley WR. Centrosome amplification and overexpression of aurora A are early events in rat mammary carcinogenesis. Cancer Res. 2002;62:4115–4122. [PubMed] [Google Scholar]
- 13.Glover DM, Leibowitz MH, McLean DA, Parry H. Mutations in aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell. 1995;81:95–105. doi: 10.1016/0092-8674(95)90374-7. [DOI] [PubMed] [Google Scholar]
- 14.Tanner MM, Grenman S, Koul A, Johannsson O, Meltzer P, Pejovic T, Borg A, Isola JJ. Frequent amplification of chromosomal region 20q12-q13 in ovarian cancer. Clin Cancer Res. 2000;6:1833–1839. [PubMed] [Google Scholar]
- 15.Tanner MM, Tirkkonen M, Kallioniemi A, Collins C, Stokke T, Karhu R, Kowbel D, Shadravan F, Hintz M, Kuo WL, et al. Increased copy number at 20q13 in breast cancer: defining the critical region and exclusion of candidate genes. Cancer Res. 1994;54:4257–4260. [PubMed] [Google Scholar]
- 16.Tanner MM, Tirkkonen M, Kallioniemi A, Holli K, Collins C, Kowbel D, Gray JW, Kallioniemi OP, Isola J. Amplification of chromosomal region 20q13 in invasive breast cancer: prognostic implications. Clin Cancer Res. 1995;1:1455–1461. [PubMed] [Google Scholar]
- 17.Lai CH, Tseng JT, Lee YC, Chen YJ, Lee JC, Lin BW, Huang TC, Liu YW, Leu TH, Chen YP, et al. Translational up-regulation of Aurora-A in EGFR-overexpressed cancer. J Cell Mol Med. 14:1520–1531. doi: 10.1111/j.1582-4934.2009.00919.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palmiter RD. Regulation of protein synthesis in chick oviduct. I. Independent regulation of ovalbumin, conalbumin, ovomucoid, and lysozyme induction. J Biol Chem. 1972;247:6450–6461. [PubMed] [Google Scholar]
- 19.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 21.Merrick WC. Cap-dependent and cap-independent translation in eukaryotic systems. Gene. 2004;332:1–11. doi: 10.1016/j.gene.2004.02.051. [DOI] [PubMed] [Google Scholar]
- 22.Jackson RJ, Hunt SL, Reynolds JE, Kaminski A. Cap-dependent and cap-independent translation: operational distinctions and mechanistic interpretations. Curr Top Microbiol Immunol. 1995;203:1–29. doi: 10.1007/978-3-642-79663-0_1. [DOI] [PubMed] [Google Scholar]
- 23.Jackson RJ, Kaminski A. Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. Rna. 1995;1:985–1000. [PMC free article] [PubMed] [Google Scholar]
- 24.Stein I, Itin A, Einat P, Skaliter R, Grossman Z, Keshet E. Translation of vascular endothelial growth factor mRNA by internal ribosome entry: implications for translation under hypoxia. Mol Cell Biol. 1998;18:3112–3119. doi: 10.1128/mcb.18.6.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holcik M, Lefebvre C, Yeh C, Chow T, Korneluk RG. A new internal-ribosome-entry-site motif potentiates XIAP-mediated cytoprotection. Nat Cell Biol. 1999;1:190–192. doi: 10.1038/11109. [DOI] [PubMed] [Google Scholar]
- 26.Pyronnet S, Pradayrol L, Sonenberg N. A cell cycle-dependent internal ribosome entry site. Mol Cell. 2000;5:607–616. doi: 10.1016/s1097-2765(00)80240-3. [DOI] [PubMed] [Google Scholar]
- 27.Stoneley M, Willis AE. Cellular internal ribosome entry segments: structures, trans-acting factors and regulation of gene expression. Oncogene. 2004;23:3200–3207. doi: 10.1038/sj.onc.1207551. [DOI] [PubMed] [Google Scholar]
- 28.Cornelis S, Bruynooghe Y, Denecker G, Van Huffel S, Tinton S, Beyaert R. Identification and characterization of a novel cell cycle-regulated internal ribosome entry site. Mol Cell. 2000;5:597–605. doi: 10.1016/s1097-2765(00)80239-7. [DOI] [PubMed] [Google Scholar]
- 29.Stoneley M, Paulin FE, Le Quesne JP, Chappell SA, Willis AE. C-Myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene. 1998;16:423–428. doi: 10.1038/sj.onc.1201763. [DOI] [PubMed] [Google Scholar]
- 30.Creancier L, Morello D, Mercier P, Prats AC. Fibroblast growth factor 2 internal ribosome entry site (IRES) activity ex vivo and in transgenic mice reveals a stringent tissue-specific regulation. J Cell Biol. 2000;150:275–281. doi: 10.1083/jcb.150.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martineau Y, Le Bec C, Monbrun L, Allo V, Chiu IM, Danos O, Moine H, Prats H, Prats AC. Internal ribosome entry site structural motifs conserved among mammalian fibroblast growth factor 1 alternatively spliced mRNAs. Mol Cell Biol. 2004;24:7622–7635. doi: 10.1128/MCB.24.17.7622-7635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van der Velden AW, Thomas AA. The role of the 5′ untranslated region of an mRNA in translation regulation during development. Int J Biochem Cell Biol. 1999;31:87–106. doi: 10.1016/s1357-2725(98)00134-4. [DOI] [PubMed] [Google Scholar]
- 33.Dobson T, Minic A, Nielsen K, Amiott E, Krushel L. Internal initiation of translation of the TrkB mRNA is mediated by multiple regions within the 5′ leader. Nucleic Acids Res. 2005;33:2929–2941. doi: 10.1093/nar/gki605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holcik M, Korneluk RG. Functional characterization of the X-linked inhibitor of apoptosis (XIAP) internal ribosome entry site element: role of La autoantigen in XIAP translation. Mol Cell Biol. 2000;20:4648–4657. doi: 10.1128/mcb.20.13.4648-4657.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mitchell SA, Spriggs KA, Coldwell MJ, Jackson RJ, Willis AE. The Apaf-1 internal ribosome entry segment attains the correct structural conformation for function via interactions with PTB and unr. Mol Cell. 2003;11:757–771. doi: 10.1016/s1097-2765(03)00093-5. [DOI] [PubMed] [Google Scholar]
- 36.Kollareddy M, Dzubak P, Zheleva D, Hajduch M. Aurora kinases: structure, functions and their association with cancer. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2008;152:27–33. doi: 10.5507/bp.2008.004. [DOI] [PubMed] [Google Scholar]
- 37.Wan XB, Long ZJ, Yan M, Xu J, Xia LP, Liu L, Zhao Y, Huang XF, Wang XR, Zhu XF, et al. Inhibition of Aurora-A suppresses epithelial-mesenchymal transition and invasion by downregulating MAPK in nasopharyngeal carcinoma cells. Carcinogenesis. 2008;29:1930–1937. doi: 10.1093/carcin/bgn176. [DOI] [PubMed] [Google Scholar]
- 38.Sukhanov S, Higashi Y, Shai SY, Itabe H, Ono K, Parthasarathy S, Delafontaine P. Novel effect of oxidized low-density lipoprotein: cellular ATP depletion via downregulation of glyceraldehyde-3-phosphate dehydrogenase. Circ Res. 2006;99:191–200. doi: 10.1161/01.RES.0000232319.02303.8c. [DOI] [PubMed] [Google Scholar]
- 39.Thomas JD, Johannes GJ. Identification of mRNAs that continue to associate with polysomes during hypoxia. RNA. 2007;13:1116–1131. doi: 10.1261/rna.534807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kozak M. Evaluation of the “scanning model” for initiation of protein synthesis in eucaryotes. Cell. 1980;22:7–8. doi: 10.1016/0092-8674(80)90148-8. [DOI] [PubMed] [Google Scholar]
- 41.Lockard RE, Lane C. Requirement for 7-methylguanosine in translation of globin mRNA in vivo. Nucleic Acids Res. 1978;5:3237–3247. doi: 10.1093/nar/5.9.3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Benedetti A, Joshi-Barve S, Rinker-Schaeffer C, Rhoads RE. Expression of antisense RNA against initiation factor eIF-4E mRNA in HeLa cells results in lengthened cell division times, diminished translation rates, and reduced levels of both eIF-4E and the p220 component of eIF-4F. Mol Cell Biol. 1991;11:5435–5445. doi: 10.1128/mcb.11.11.5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duncan R, Milburn SC, Hershey JW. Regulated phosphorylation and low abundance of HeLa cell initiation factor eIF-4F suggest a role in translational control. Heat shock effects on eIF-4F. J Biol Chem. 1987;262:380–388. [PubMed] [Google Scholar]
- 44.Hiremath LS, Webb NR, Rhoads RE. Immunological detection of the messenger RNA cap-binding protein. J Biol Chem. 1985;260:7843–7849. [PubMed] [Google Scholar]
- 45.Avdulov S, Li S, Michalek V, Burrichter D, Peterson M, Perlman DM, Manivel JC, Sonenberg N, Yee D, Bitterman PB, et al. Activation of translation complex eIF4F is essential for the genesis and maintenance of the malignant phenotype in human mammary epithelial cells. Cancer Cell. 2004;5:553–563. doi: 10.1016/j.ccr.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 46.De Benedetti A, Graff JR. eIF-4E expression and its role in malignancies and metastases. Oncogene. 2004;23:3189–3199. doi: 10.1038/sj.onc.1207545. [DOI] [PubMed] [Google Scholar]
- 47.Lazaris-Karatzas A, Smith MR, Frederickson RM, Jaramillo ML, Liu YL, Kung HF, Sonenberg N. Ras mediates translation initiation factor 4E-induced malignant transformation. Genes Dev. 1992;6:1631–1642. doi: 10.1101/gad.6.9.1631. [DOI] [PubMed] [Google Scholar]
- 48.Ramirez-Valle F, Braunstein S, Zavadil J, Formenti SC, Schneider RJ. eIF4GI links nutrient sensing by mTOR to cell proliferation and inhibition of autophagy. J Cell Biol. 2008;181:293–307. doi: 10.1083/jcb.200710215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haghighat A, Sonenberg N. eIF4G dramatically enhances the binding of eIF4E to the mRNA 5′-cap structure. J Biol Chem. 1997;272:21677–21680. doi: 10.1074/jbc.272.35.21677. [DOI] [PubMed] [Google Scholar]
- 50.Dobson T, Kube E, Timmerman S, Krushel LA. Identifying intrinsic and extrinsic determinants that regulate internal initiation of translation mediated by the FMR1 5′ leader. BMC Mol Biol. 2008;9:89. doi: 10.1186/1471-2199-9-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gingras AC, Gygi SP, Raught B, Polakiewicz RD, Abraham RT, Hoekstra MF, Aebersold R, Sonenberg N. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 1999;13:1422–1437. doi: 10.1101/gad.13.11.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jan E, Sarnow P. Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J Mol Biol. 2002;324:889–902. doi: 10.1016/s0022-2836(02)01099-9. [DOI] [PubMed] [Google Scholar]
- 53.Jang SK, Davies MV, Kaufman RJ, Wimmer E. Initiation of protein synthesis by internal entry of ribosomes into the 5′ nontranslated region of encephalomyocarditis virus RNA in vivo. J Virol. 1989;63:1651–1660. doi: 10.1128/jvi.63.4.1651-1660.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Eden ME, Byrd MP, Sherrill KW, Lloyd RE. Demonstrating internal ribosome entry sites in eukaryotic mRNAs using stringent RNA test procedures. RNA. 2004;10:720–730. doi: 10.1261/rna.5225204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thompson SR. So you want to know if your message has an IRES? Wiley Interdiscip Rev RNA. doi: 10.1002/wrna.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lewis SM, Holcik M. For IRES trans-acting factors, it is all about location. Oncogene. 2008;27:1033–1035. doi: 10.1038/sj.onc.1210777. [DOI] [PubMed] [Google Scholar]
- 57.Tinton SA, Schepens B, Bruynooghe Y, Beyaert R, Cornelis S. Regulation of the cell-cycle-dependent internal ribosome entry site of the PITSLRE protein kinase: roles of Unr (upstream of N-ras) protein and phosphorylated translation initiation factor eIF-2alpha. Biochem J. 2005;385:155–163. doi: 10.1042/BJ20040963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bergamini G, Preiss T, Hentze MW. Picornavirus IRESes and the poly(A) tail jointly promote cap-independent translation in a mammalian cell-free system. RNA. 2000;6:1781–1790. doi: 10.1017/s1355838200001679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dehlin E, Wormington M, Korner CG, Wahle E. Cap-dependent deadenylation of mRNA. EMBO J. 2000;19:1079–1086. doi: 10.1093/emboj/19.5.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chiang PW, Carpenter LE, Hagerman PJ. The 5′-untranslated region of the FMR1 message facilitates translation by internal ribosome entry. J Biol Chem. 2001;276:37916–37921. doi: 10.1074/jbc.M105584200. [DOI] [PubMed] [Google Scholar]
- 61.Band V, editor. Tumor progression in breast cancer. Humana Press; Totowa (NJ): 1991. [Google Scholar]
- 62.Band V, Sager R. Distinctive traits of normal and tumor-derived human mammary epithelial cells expressed in a medium that supports long-term growth of both cell types. Proc Natl Acad Sci U S A. 1989;86:1249–1253. doi: 10.1073/pnas.86.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Band V, Zajchowski D, Swisshelm K, Trask D, Kulesa V, Cohen C, Connolly J, Sager R. Tumor progression in four mammary epithelial cell lines derived from the same patient. Cancer Res. 1990;50:7351–7357. [PubMed] [Google Scholar]
- 64.Hata T, Furukawa T, Sunamura M, Egawa S, Motoi F, Ohmura N, Marumoto T, Saya H, Horii A. RNA interference targeting aurora kinase a suppresses tumor growth and enhances the taxane chemosensitivity in human pancreatic cancer cells. Cancer Res. 2005;65:2899–2905. doi: 10.1158/0008-5472.CAN-04-3981. [DOI] [PubMed] [Google Scholar]
- 65.Neben K, Korshunov A, Benner A, Wrobel G, Hahn M, Kokocinski F, Golanov A, Joos S, Lichter P. Microarray-based screening for molecular markers in medulloblastoma revealed STK15 as independent predictor for survival. Cancer Res. 2004;64:3103–3111. doi: 10.1158/0008-5472.can-03-3968. [DOI] [PubMed] [Google Scholar]
- 66.Sen S, Zhou H, White RA. A putative serine/threonine kinase encoding gene BTAK on chromosome 20q13 is amplified and overexpressed in human breast cancer cell lines. Oncogene. 1997;14:2195–2200. doi: 10.1038/sj.onc.1201065. [DOI] [PubMed] [Google Scholar]
- 67.Tseng YS, Lee JC, Huang CY, Liu HS. Aurora-A overexpression enhances cell-aggregation of Haras transformants through the MEK/ERK signaling pathway. BMC Cancer. 2009;9:435. doi: 10.1186/1471-2407-9-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kozak M. A second look at cellular mRNA sequences said to function as internal ribosome entry sites. Nucleic Acids Res. 2005;33:6593–6602. doi: 10.1093/nar/gki958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Holcik M. Targeting translation for treatment of cancer--a novel role for IRES? Curr Cancer Drug Targets. 2004;4:299–311. doi: 10.2174/1568009043333005. [DOI] [PubMed] [Google Scholar]
- 70.Coldwell MJ, Mitchell SA, Stoneley M, MacFarlane M, Willis AE. Initiation of Apaf-1 translation by internal ribosome entry. Oncogene. 2000;19:899–905. doi: 10.1038/sj.onc.1203407. [DOI] [PubMed] [Google Scholar]
- 71.Sehgal A, Briggs J, Rinehart-Kim J, Basso J, Bos TJ. The chicken c-Jun 5′ untranslated region directs translation by internal initiation. Oncogene. 2000;19:2836–2845. doi: 10.1038/sj.onc.1203601. [DOI] [PubMed] [Google Scholar]
- 72.Sherrill KW, Byrd MP, Van Eden ME, Lloyd RE. BCL-2 translation is mediated via internal ribosome entry during cell stress. J Biol Chem. 2004;279:29066–29074. doi: 10.1074/jbc.M402727200. [DOI] [PubMed] [Google Scholar]
- 73.Yang DQ, Halaby MJ, Zhang Y. The identification of an internal ribosomal entry site in the 5′-untranslated region of p53 mRNA provides a novel mechanism for the regulation of its translation following DNA damage. Oncogene. 2006;25:4613–4619. doi: 10.1038/sj.onc.1209483. [DOI] [PubMed] [Google Scholar]
- 74.Bellodi C, Kopmar N, Ruggero D. Deregulation of oncogene-induced senescence and p53 translational control in X-linked dyskeratosis congenita. EMBO J. 29:1865–1876. doi: 10.1038/emboj.2010.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Montanaro L, Calienni M, Bertoni S, Rocchi L, Sansone P, Storci G, Santini D, Ceccarelli C, Taffurelli M, Carnicelli D, et al. Novel dyskerin-mediated mechanism of p53 inactivation through defective mRNA translation. Cancer Res. 70:4767–4777. doi: 10.1158/0008-5472.CAN-09-4024. [DOI] [PubMed] [Google Scholar]
- 76.Yoon A, Peng G, Brandenburger Y, Zollo O, Xu W, Rego E, Ruggero D. Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science. 2006;312:902–906. doi: 10.1126/science.1123835. [DOI] [PubMed] [Google Scholar]
- 77.Braunstein S, Karpisheva K, Pola C, Goldberg J, Hochman T, Yee H, Cangiarella J, Arju R, Formenti SC, Schneider RJ. A hypoxia-controlled cap-dependent to cap-independent translation switch in breast cancer. Mol Cell. 2007;28:501–512. doi: 10.1016/j.molcel.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 78.Filbin ME, Kieft JS. Toward a structural understanding of IRES RNA function. Curr Opin Struct Biol. 2009;19:267–276. doi: 10.1016/j.sbi.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Belsham GJ, Sonenberg N. Picornavirus RNA translation: roles for cellular proteins. Trends Microbiol. 2000;8:330–335. doi: 10.1016/s0966-842x(00)01788-1. [DOI] [PubMed] [Google Scholar]
- 80.Pilipenko EV, Blinov VM, Romanova LI, Sinyakov AN, Maslova SV, Agol VI. Conserved structural domains in the 5′-untranslated region of picornaviral genomes: an analysis of the segment controlling translation and neurovirulence. Virology. 1989;168:201–209. doi: 10.1016/0042-6822(89)90259-6. [DOI] [PubMed] [Google Scholar]
- 81.Vagner S, Galy B, Pyronnet S. Irresistible IRES. Attracting the translation machinery to internal ribosome entry sites. EMBO Rep. 2001;2:893–898. doi: 10.1093/embo-reports/kve208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Beaudoin ME, Poirel VJ, Krushel LA. Regulating amyloid precursor protein synthesis through an internal ribosomal entry site. Nucleic Acids Res. 2008;36:6835–6847. doi: 10.1093/nar/gkn792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chappell SA, Edelman GM, Mauro VP. A 9-nt segment of a cellular mRNA can function as an internal ribosome entry site (IRES) and when present in linked multiple copies greatly enhances IRES activity. Proc Natl Acad Sci U S A. 2000;97:1536–1541. doi: 10.1073/pnas.97.4.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jang GM, Leong LE, Hoang LT, Wang PH, Gutman GA, Semler BL. Structurally distinct elements mediate internal ribosome entry within the 5′-noncoding region of a voltage-gated potassium channel mRNA. J Biol Chem. 2004;279:47419–47430. doi: 10.1074/jbc.M405885200. [DOI] [PubMed] [Google Scholar]
- 85.Ali N, Siddiqui A. The La antigen binds 5′ noncoding region of the hepatitis C virus RNA in the context of the initiator AUG codon and stimulates internal ribosome entry site-mediated translation. Proc Natl Acad Sci U S A. 1997;94:2249–2254. doi: 10.1073/pnas.94.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Borovjagin A, Pestova T, Shatsky I. Pyrimidine tract binding protein strongly stimulates in vitro encephalomyocarditis virus RNA translation at the level of preinitiation complex formation. FEBS Lett. 1994;351:299–302. doi: 10.1016/0014-5793(94)00848-5. [DOI] [PubMed] [Google Scholar]
- 87.Pfingsten JS, Costantino DA, Kieft JS. Structural basis for ribosome recruitment and manipulation by a viral IRES RNA. Science. 2006;314:1450–1454. doi: 10.1126/science.1133281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hellen CU, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- 89.Gu L, Zhu N, Zhang H, Durden DL, Feng Y, Zhou M. Regulation of XIAP translation and induction by MDM2 following irradiation. Cancer Cell. 2009;15:363–375. doi: 10.1016/j.ccr.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Halaby MJ, Hibma JC, He J, Yang DQ. ATM protein kinase mediates full activation of Akt and regulates glucose transporter 4 translocation by insulin in muscle cells. Cell Signal. 2008;20:1555–1563. doi: 10.1016/j.cellsig.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 91.Ohno S, Shibayama M, Sato M, Tokunaga A, Yoshida N. Polypyrimidine tract-binding protein regulates the cell cycle through IRES-dependent translation of CDK11(p58) in mouse embryonic stem cells. Cell Cycle. 10:3706–3713. doi: 10.4161/cc.10.21.17903. [DOI] [PubMed] [Google Scholar]
- 92.Lee EC, Frolov A, Li R, Ayala G, Greenberg NM. Targeting Aurora kinases for the treatment of prostate cancer. Cancer Res. 2006;66:4996–5002. doi: 10.1158/0008-5472.CAN-05-2796. [DOI] [PubMed] [Google Scholar]
- 93.Kitzen JJ, de Jonge MJ, Verweij J. Aurora kinase inhibitors. Crit Rev Oncol Hematol. 2009 doi: 10.1016/j.critrevonc.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 94.Mountzios G, Terpos E, Dimopoulos MA. Aurora kinases as targets for cancer therapy. Cancer Treat Rev. 2008;34:175–182. doi: 10.1016/j.ctrv.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 95.Dutertre S, Descamps S, Prigent C. On the role of aurora-A in centrosome function. Oncogene. 2002;21:6175–6183. doi: 10.1038/sj.onc.1205775. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.