Fig. 3.
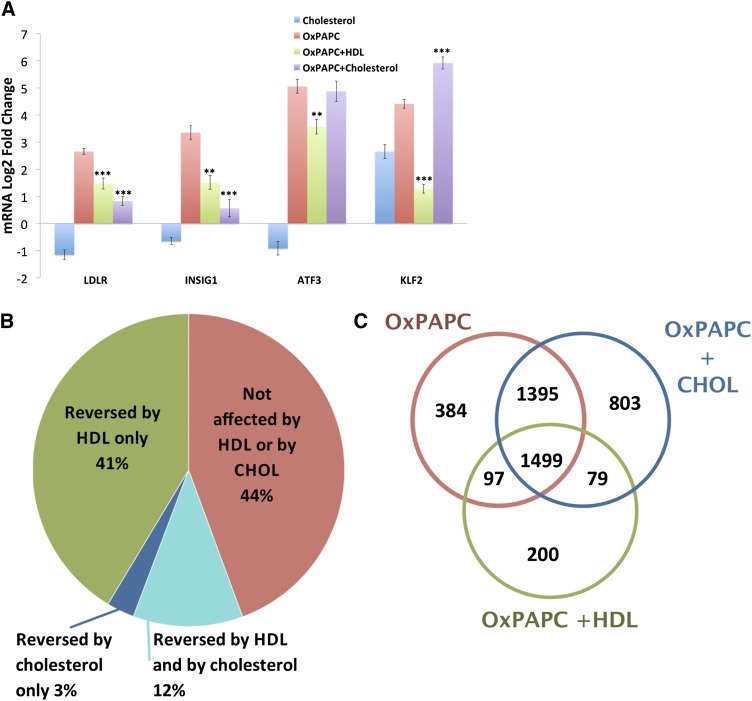
Cholesterol loading mimics some aspects of the HDL response. To determine the extent to which HDL’s effect on OxPAPC signaling in HAECs is mediated by cholesterol repletion, we compared the effect of cholesterol loading using cholesterol-cyclodextrin with that of HDL. HAECs were pretreated for 1 h with media containing 1% FBS with or without HDL (50 μg/ml) or cholesterol-cyclodextrin (20 μg/ml). Media or OxPAPC was then added to a final concentration of 50 μg/ml. A: After a 4 h treatment, mRNA was isolated and quantified by qPCR. The expression of SREBP target genes (LDLR and INSIG1) was induced by OxPAPC and similarly affected by HDL and cholesterol-cyclodextrin cotreatment. In contrast, HDL inhibited the OxPAPC-mediated induction of ATF3 and KLF2, whereas cholesterol-cyclodextrin had no effect and enhanced the upregulation of ATF3 and KLF2, respectively. Data are presented as average log2-fold change of triplicates ± SEM. * P < 0.05, ** P < 0.01, *** P < 0.001 for comparison with OxPAPC log2-fold changes. P values defined by unpaired Student’s t-test. B, C: To further characterize the similarity between the effects of HDL and cholesterol-cyclodextrin on OxPAPC signaling in HAECs, we repeated the treatment described previously on three additional HAEC donors and quantified global mRNA expression by microarray. B: Pie chart summarizing the overlap between the effects of HDL and cholesterol-cyclodextrin on genes affected by OxPAPC. Consistent with our previous experiment, HDL reversed a majority (53%) of genes affected by OxPAPC. In contrast, cholesterol-cyclodextrin reversed only 23% of genes reversed by HDL. Only 3% of genes affected by OxPAPC were reversed by cholesterol but not by HDL. C: Venn diagram, comparing cholesterol treatment versus HDL treatment. Cholesterol-cyclodextrin, when combined with OxPAPC, had synergistic effects on gene expression as reflected by the number of genes (i.e., 803) affected only in presence of both. OxPAPC and HDL, however, had a limited synergistic effect on gene expression.