Abstract
Lysophosphatidic acids (LPAs) are biologically active signaling molecules involved in the regulation of many cellular processes and have been implicated as potential mediators of fibroblast recruitment to the pulmonary airspace, pointing to possible involvement of LPA in the pathology of pulmonary fibrosis. LPAs have been measured in various biological matrices and many challenges involved with their analyses have been documented. However, little published information is available describing LPA levels in human bronchoalveolar lavage fluid (BALF). We therefore conducted detailed investigations into the effects of extensive sample handling and sample preparation conditions on LPA levels in human BALF. Further, targeted lipid profiling of human BALF and plasma identified the most abundant lysophospholipids likely to interfere with LPA measurements. We present the findings from these investigations, highlighting the importance of well-controlled sample handling for the accurate quantitation of LPA. Further, we show that chromatographic separation of individual LPA species from their corresponding lysophospholipid species is critical to avoid reporting artificially elevated levels. The optimized sample preparation and LC/MS/MS method was qualified using a stable isotope-labeled LPA as a surrogate calibrant and used to determine LPA levels in human BALF and plasma from a Phase 0 clinical study comparing idiopathic pulmonary fibrosis patients to healthy controls.
Keywords: bronchoalveolar lavage fluid, human plasma, stability, idiopathic pulmonary fibrosis
Lysophosphatidic acids (LPAs) are intermediates in phospholipid synthesis and important mediators of many cellular processes including growth, motility, and differentiation. LPA have also been implicated in a variety of diseases such as atherosclerosis, respiratory disorders, gynecological cancer, and fibrosis (1–3). Many of the pleiotropic growth factor-type effects and signaling processes of LPA are mediated through G protein-coupled receptors, the most studied of which have been designated LPA1-LPA5 receptors (4, 5). LPA1 receptor, in particular, has been linked to fibrosis (2, 3). Studies by Tager et al. (3) suggested that LPAs, signaling through LPA1 receptor, are chemotactic for fibroblast recruitment, and that LPA levels are higher in the bronchoalveolar lavage fluid (BALF) of patients with idiopathic pulmonary fibrosis (IPF) compared with controls. IPF is a chronic and progressive disease with high mortality and a median survival of 2 to 4 years and there are currently no approved therapies in the US other than transplant (6). The potential involvement of LPA in lung fibrosis suggests that targeting LPA1 receptor for pharmaceutical intervention may hold promise as a treatment for IPF.
The potential for use of LPA as biomarkers for diseases such as ovarian cancer, as well as their potential involvement in fibrotic disease, has resulted in the measurement of LPA in plasma and, more recently, BALF using a variety of analytical techniques (for a recent review, see Jesionowska et al.) (7). As reported by Smyth et al. (8), the levels of LPA in human plasma can differ by as much as 10-fold depending on the method of analysis and the individual laboratory. It is notable that LPA levels in human plasma measured using enzymatic or radioenzymatic assays have been reported in the 80 nM range, while LPA levels measured using ESI-MS are much higher and vary among laboratories (600 to >1200 nM in healthy human plasma) (8–17). The number of documented measurements of LPA in BALF is much more limited, and it is therefore difficult to comment on the variability of LPA levels reported between laboratories. BALF LPA levels reported to date have been measured by LC/MS and range from approximately 0.5 to <1000 nM in human BALF (following allergen challenge) and approximately 5-20 nM in human BALF from healthy or IPF patients (3, 18, 19).
It is perhaps not surprising that such a variation in reported LPA levels exists, since it has been documented that LPA can be generated ex vivo post-sample collection through several analytical, chemical, and biological processes, making accurate analytical measurement of LPA in biological fluids challenging. Zhao et al. were the first to report that LPA can be generated from other lysophospholipids, such as lysophosphatidylcholine (LPC), in the ionization source during MS analysis (20). To overcome this issue, interfering lysophospholipids must be either chromatographically separated from LPA or selectively removed during the sample preparation process. Typically, a TLC or liquid-liquid extraction (LLE) clean-up step is used followed by either direct flow injection analysis or LC/MS (16). While Zhao et al. focused on chromatographic separation of LPA from LPC (following a chloroform based LLE step) we found (through lipid profiling described in a later section) significant levels of various lysophospholipids, particularly lysophosphatidylserine (LPS), in human BALF. As Zhao described, we also found that LPS, as well as other lysophospholipids, convert to LPA by in-source fragmentation, a common phenomenon in mass spectrometric measurements. Source fragmentation can sometimes be minimized by optimizing ionization conditions but often cannot be completely eliminated and can present a significant challenge when a more endogenously abundant species (i.e., LPC or LPS) fragments to form the species of interest (i.e., LPA) that are much less abundant. This was true in our case where we found that, because of the low levels of LPA in human BALF, it was necessary to chromatographically separate LPA from several lysophospholipid species (found in BALF and plasma in uM concentration) to avoid reporting artificially high levels of LPA.
Besides MS-specific generation of LPA from lysophospholipids, LPAs are also formed through enzymatic or chemical conversion during sample handling and the sample preparation process. LPA can be generated enzymatically in plasma via the action of lysophospholipase D (lysoPLD) on LPC. The lysoPLD activity in plasma is presumed to be due primarily to autotaxin, a multifunctional phosphodiesterase (21–24). Human plasma and serum LPA have been shown to increase in a time-dependent manner upon incubation at 25 or 37°C, indicating that lysoPLD activity persists ex vivo (11, 13, 24–26). Chemical conversion of lysophospholipid such as LPC to LPA can also occur in the presence of strong acid (26). The most commonly reported sample preparation method for LPA extraction from biological fluids involves LLE with acidification (3, 12, 14, 15, 18, 19, 27–29). Scherer et al. showed that under strong acidic conditions, LPC degrades to LPA in the presence of plasma, and the total LPA concentration in human plasma was almost 10-fold higher in plasma extracted under acidic conditions versus plasma extracted with butanol (26). These findings suggest that differences in sample handling could explain some of the differences in reported endogenous LPA levels in the literature.
Our ultimate objective (based on literature reports citing LPA as potential biomarkers of various disease states) was to measure the concentration of LPA in plasma and BALF from a Phase 0 study comparing healthy controls to IPF patients. Given the knowledge that LPA levels in biological fluids can 1) continue to increase post-sample collection due to persistent enzymatic activity, 2) be generated from degradation of other endogenous lysophospholipids, and 3) be formed in the MS source, our goal was to develop an accurate LC/MS/MS method for the quantitation of a panel of LPAs in human plasma and BALF from a Phase 0 study. Because the suspected sources of ex vivo LPA generation (i.e., other phosphoplipids) were expected at varying basal levels between patients and could, therefore, convert to LPA to different degrees, the need for accurate determination of “true” LPA levels was paramount. Given the shortage of information describing LPA in human BALF, we first performed targeted lipid profiling to identify the most abundant LPA and lysophospholipid species. We then characterized the gas phase conversion potential of the most abundant lysophospholipids to the corresponding LPA using synthetic standards and determined that chromatographic separation of LPA from several lysophospholipid species, in particular LPS (which proved very difficult to separate from LPA), was needed to ensure accurate quantitation. Following chromatographic, MS, and sample preparation condition optimization, detailed stability studies were conducted and the method was qualified using an in-house synthetic stable isotope-labeled LPA as a surrogate calibrant for endogenous LPA. LPA levels were then determined in Phase 0 plasma and BALF samples.
We present a sensitive and extremely selective method for the determination of LPA in biological fluids. We also summarize the LPA and lysophospholipid lipid profiling patterns in human BALF and the stability characterization of endogenous LPA in plasma and BALF, showing that sample handling can alter not only the LPA concentration reported, but the overall lysophospholipid profile as well.
MATERIALS AND METHODS
Materials
Water, methanol, 1- butanol, isopropanol (all Chromasolv® Plus for HPLC), formic acid (> 95%), ammonium acetate, citric acid, sodium phosphate dibasic, calcium chloride, sodium deoxycholate, Tris buffer, and phospholipase A2 from bovine pancreas (≥20 units/mg protein) were from Sigma-Aldrich (St. Louis, MO). 16:0-LPA, 17:0-LPA, 18:0-LPA, 18:1-LPA, 20:4-LPA, C18:0-LPS, d35-C18:0-LPC, and 1,2-didocosahexaenoyl-sn-glycero-3-phosphate (22:6-PA) were from Avanti Polar Lipids (Alabaster, AL). Human plasma (K2EDTA, gender pooled) and BALF (gender pooled) were obtained from Bioreclamation (Liverpool, NY). d5-2-hydroxy-3-(phosphonooxy)propyl palmitate LPA (d5-C16:0-LPA) was synthesized in-house as described below.
Synthesis of d5-C16:0-LPA standard
The d5-C16:0-LPA was prepared using a three-step synthetic route from (S)d5-glycidol. Phosphorylation of (S)d5-glycidol using di-tertbutyl-N,N-diisopropyl phosphoramidite and subsequent oxidation with m-CPBA produced (R)-di-tert-butyl-phosphoyl-d5-glycidol. Regioselective opening of the epoxide with cesium palmitate produced the d5-di-tert-butyl-protected lysoalkyl phosphatidic acid. Deprotection with trifluoroacetic acid in dichloromethane yielded the d5-2-hydroxy-3-(phosphonooxy)propyl palmitate LPA. The chemical purity was determined to be 98.3% by HPLC/UV. Isotopic purity was determined to be 90% by high resolution MS. Both chemical and isotopic purity were taken into consideration when making stock solutions.
Synthesis of 22:6-LPA (for retention time confirmation)
Synthesis of 22:6-LPA was accomplished using 1,2-didocosahexaenoyl-sn-glycero-3-phosphate (22:6-PA) and phosphoplipase A2 as described in the literature (30). Briefly, 1 mg of 22:6-PA was dried in a round bottom glass cylinder under a nitrogen gas stream. To the residue were added 2 ml of buffer (50 mM Tris pH 7.5, 10 mM CaCl2, 1 mM sodium deoxycholate) and 30 units of phospholipase A2. The reaction was vortexed then stirred overnight (∼16 h) under a constant nitrogen stream. The sample was taken to dryness, reconstituted in methanol, and split in two. One portion was used for characterization by high resolution LC/MS (Thermo Accela Q-Exactive mass spectrometer) for verification of the expected elemental composition (within 3 ppm); the second portion was analyzed by LC/MS/MS as described below. Overall yield was not determined.
LC/MS/MS conditions
The LC/MS/MS system consisted of an HTC Pal autosampler coupled to a Leap Flux uHPLC pump (Leap Technologies, Carrboro, NC) and a Sciex (Framingham, MA) API5500 mass spectrometer fitted with a TurboIonSpray® probe. LC was performed on a Phenomenex (Torrance, CA) Kinetix C18 2.1 × 100 mM 1.7 µm column (room temperature) using a flow rate of 0.35 ml/min and a mobile phase consisting of solvent A = 0.1% formic acid/5 mM ammonium acetate in 95:5 water:methanol and solvent B = 0.1% formic acid/5 mM ammonium acetate in 5:95 water:methanol. Gradient conditions were as follows: hold 30% B 0.5 min, 30–80% B in 0.5 min, hold 1 min, 80–100% B in 7 min, hold 3 min, reequilibrate 1 min to 30% B; the first 0.6 min of each run was diverted to waste. Autosampler wash solvents consisted of 0.2% formic acid in 80:20 water:methanol (wash 1) and 0.2% formic acid in 1:1 methanol:isopropanol (wash 2); the injection volume was 10 µl. Optimized MS conditions (ESI, negative mode) were as follows: curtain gas 35, GS1 55, GS2 45, CAD gas 7, source temperature 500°C, IS voltage -4500V, DP -40, EP -4, CE -40, and CXP -11. As previously mentioned, these conditions were optimized to minimize source fragmentation via loss of the polar head groups from various lysophospholipids to form identical acyl chain LPA. LPAs, LPSs, LPIs (lysophosphatidylinositols) and LPGs (lysophosphatidylglycerols) were monitored as the transition of their (M-H)- ion to m/z 152.9. LPC transitions were based on fragmentation of the (M-44)- ion to the corresponding acylated fragment ion. MRM transitions monitored for the primary lysophospholipid of interest were: d5-C16:0-LPA 414.2→157.9, C16:0-LPA 409.2→152.9, C16:1-LPA 407.2→152.9, C17:0-LPA (IS) 423.2→152.9, C18:0-LPA 437.2→152.9, C18:1-LPA 435.2→152.9, C18:2-LPA 433.2→152.9, C20:4-LPA 457.2→152.9, C22:6-LPA 481.2→152.9, C18:0-LPS 524.3→152.9.
Sample preparation
LLE using butanol was based on methods described in the literature (11). Briefly, 200 µl of human plasma or BALF (in a 2 ml polypropylene microcentrifuge tube) was spiked with 10 µl of internal standard (IS) (C17:0-LPA) working solution (in methanol) followed by 200 ul of extraction buffer (30 mM citrate/40 mM sodium phosphate in water) and 600 µl of butanol. The IS used, C17:0-LPA, was not detected as an endogenous component of human plasma or BALF. Samples were mixed by vortexing vigorously for 2 min, then centrifuged in a tabletop microcentrifuge at top speed for 10 min. The top organic layer was transferred to a 96-well plate and dried under heated (∼50°C) nitrogen. The residues were reconstituted in 200 µl of methanol, vortexed vigorously for 2 min then centrifuged for 10 min at 4,000 rpm in a refrigerated centrifuge fitted with a 96-well plate rotor. The supernatant was transferred to a clean plate and the samples were analyzed by LC/MS/MS as described above.
Quantitation
LPA stock solutions were prepared at 0.2 mg/ml in methanol and stored at −20°C. LPAs of interest were quantitated using d5-C16:0-LPA as a surrogate calibrant. D5-C16:0-LPA calibration curves were generated using C17:0-LPA as an IS; the typical calibration range was 0.1–100 ng/ml (0.24–241 nM, based on 200 µl of matrix), fitting was quadratic 1/x*x. Following processing of all data using Analyst (v1.5.1), GraphPad Prism 5 was used to generate d5-C16:0-LPA calibration curves and to interpolate the concentration of LPA of interest in the human plasma or BALF samples.
Stability testing
Bench-top stability experiments were conducted by incubating human plasma or BALF on ice and removing 200 µl time-points at 0, 0.5, 1, and 4 h. Time-points were immediately processed using the LLE protocol described above.
Freeze-thaw stability experiments were conducted by subjecting human plasma or BALF to three freeze-thaw cycles with at least one 24 h cycle. At the end of the stability experiment the samples were prepared as described above.
Phase 0 subjects
A single center cross-sectional study was conducted in order to compare levels of biomarkers between IPF and control subjects. Subjects with IPF (n = 4) strictly defined according to (American Thoracic Society criteria and with FEV1/FVC >0.75 (FEV1 = forced exploratory volume in 1 s, FVC = forced vital capacity) and control subjects (n = 5–6) were recruited at the Duke University Clinical Research Unit. The clinical protocol was approved by the Duke University Medical Center Copernicus IRB, and informed consent was obtained from all patients involved. EDTA-anti-coagulated blood was drawn for plasma, and subjects underwent identical bronchoscopy and BALF collection procedures. This noninterventional study is referred to as ‘Phase 0’ throughout the article.
RESULTS
Targeted lipid profiling of human plasma and BALF
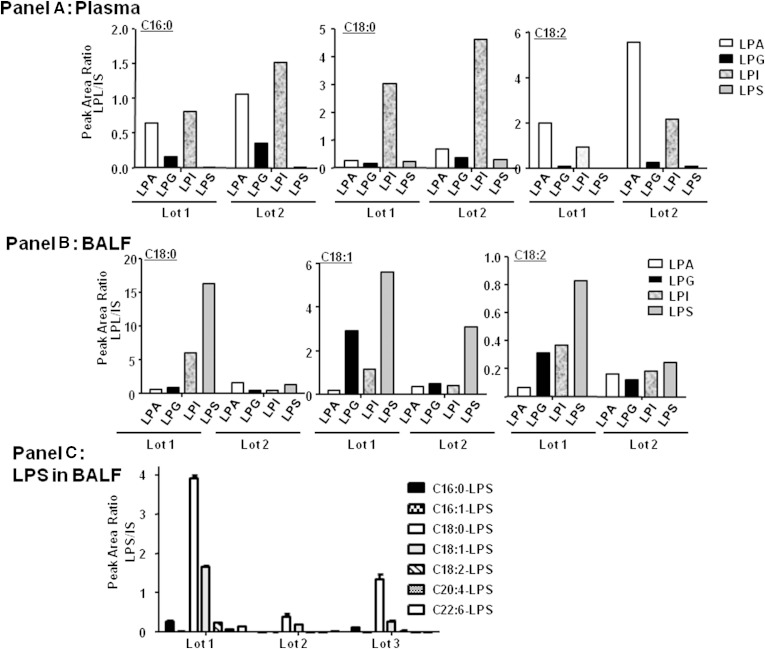
Considering the limited characterization of the lysophospholipid population of human BALF, we began by characterizing the general lysophospholipid content of BALF to determine which species were most likely to interfere with LPA analysis. Although more information is available for the human plasma lipid profile, we conducted all studies in both matrices to allow direct comparisons where applicable. Qualitative targeted lipid profiling was performed on human plasma and human BALF liquid-liquid extracts using high resolution MS, focusing on the comparison of various lysophospholipid species in each biological fluid. The ionization response of lipids is known to vary depending on the acyl chain length, degree of unsaturation, head group, concentration, mobile phase composition, and instrument parameters (31–33). However, because our focus was to identify which lysophospholipid had the greatest potential to interfere with LPA analysis, a qualitative comparison of lysophospholipid levels in human plasma and BALF under the conditions to be used for LPA analysis was sufficient (31, 33). As has been reported elsewhere, the majority of the plasma lysophospholipid pool consists of LPCs followed closely by lysophosphatidylethanolamines, which have been documented to be in the µM range (34–36). Qualitatively, other lysophospholipid classes (LPIs, LPGs, LPSs, and LPAs) were present at much lower levels in plasma compared with LPCs. Figure 1A shows the qualitative lysophospholipid profile (excluding highly abundant LPCs and lysophosphatidylethanolamines) of human plasma from two gender pooled plasma lots. These lots were purchased separately and were meant to represent the potential variation in plasma lysophospholipid that might be expected. For simplification of the figure, C16:0-, C18:0-, and C18:2- were chosen as representative acyl substituents. In general, the lysophospholipid distribution profile was similar between lots of plasma. BALF, however, showed more variation in lysophospholipid distribution and relative abundance from lot to lot (Fig. 1B). Given the anticipated low levels of LPA in human BALF, we were especially concerned about potentially interfering lysophospholipid. One consistent observation was that LPS seemed to be present as the most relatively abundant species in the lots of human BALF tested. Focusing on LPS (Fig. 1C), of the three human BALF lots (each BALF lot was gender pooled) tested, C18:0-LPS and C18:1-LPS were the most abundant. These results suggested that chromatographic separation of LPA from other lysophospholipids was necessary to avoid reporting artificially high levels of LPA in the Phase 0 BALF samples, which would contain an uncharacterized lysophospholipid population.
Fig. 1.
Targeted lipid profiling of human plasma/BALF. Human plasma (A) or BALF (B) was extracted and analyzed by LC/MS/MS as described under Materials and Methods. Extracted ion chromatograms for different LPSs are shown in C.
Chromatographic method development
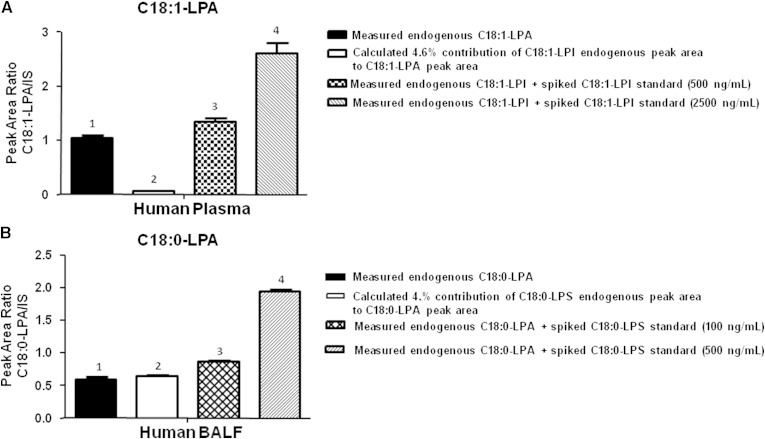
Purchased LPG, LPI, LPS, and LPC standards were used to determine the approximate in-source percent conversion to LPA. The in-source conversion potential varied from standard to standard but ranged from approximately 0.1% to 4% (data not shown). Manipulation of MS source parameters did not eliminate or drastically affect in-source conversion potential. The probability of each lysophospholipid class to convert to LPA in the source was taken into consideration when developing and testing the chromatographic separation conditions. For example, C18:1-LPI showed ∼4.6% in-source conversion to C18:1-LPA; because LPIs were present at relatively similar levels to LPA in plasma (Fig. 1A), LPI chromatographic separation from LPA was considered essential. Likewise, C18:0-LPS showed ∼4% conversion to C18:0-LPA and given the relatively high concentration of LPS versus LPA in BALF (Fig. 1B, C), chromatographic separation of LPS from LPA in BALF was considered necessary.
Using a previously developed LC method that effectively separated major lipid classes from one another (i.e., phosphatidylcholines, triglycerides, fatty acids, lysophospholipids) but did not necessarily separate individual lysophospholipid species, we tested the effect of spiking various lysophospholipid standards into human plasma and BALF on measured endogenous LPA levels. Figure 2A shows the results obtained following spiking of human plasma extract with either 500 or 2,500 ng/ml of purchased C18:1-LPI standard (unspiked plasma was used for baseline determination, bar 1). In the baseline sample, the peak area of endogenous C18:1-LPI was determined and multiplied by 4.6% to obtain the theoretical in-source conversion that could contribute to C18:1-LPA (Fig. 2A, bar 2). As shown in bar 1 and 2 of Fig. 2A, the theoretical contribution of endogenous C18:1-LPI to C18:1-LPA is relatively minor (in this particular plasma sample). However, if additional C18:1-LPI was spiked into the sample, the artificial enhancement of measured C18:1-LPA (due to insufficient separation of the LPA and LPS peaks) is obvious (Fig. 2A, bars 3 and 4). A similar experiment was conducted with human BALF (Fig. 2B) spiked with 100 or 500 ng/ml of C18:0-LPS standard. In this sample, the peak area of endogenous C18:0-LPA (Fig. 2B, bar 1) could be theoretically entirely due to artificial contribution from coeluting endogenous C18:0-LPS (Fig. 2B, bar 2). Further elevation of C18:0-LPA signal is evident following spiking of the BALF with C18:0-LPS standard (Fig. 2B, bars 3 and 4).
Fig. 2.
Demonstration of LPS contribution to LPA in BALF using low resolution chromatography. Human plasma was spiked with C18:1-LPI (A) at 500 or 2500 ng/ml and human BALF was spiked with C18:0-LPS (B) at 100 or 500 ng/ml and extracted and analyzed by LC/MS/MS as described in Materials and Methods. C18:1-LPA was monitored to show contribution of the spiked components to the LPA channel.
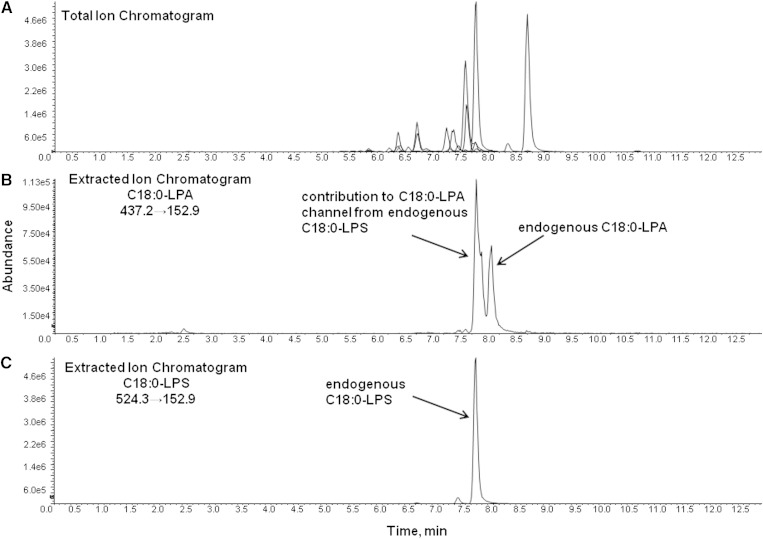
LC conditions were optimized to separate individual LPA from their corresponding lysophospholipid counterparts (i.e., C18:0-LPA from C18:0-LPC, C18:0-LPS, C18:0-LPI, etc.); separation of LPA from LPS proved to be the most challenging. Sufficient separation was eventually achieved on a Phenomenex Kinetix C18 column using a mobile phase of methanol and aqueous ammonium acetate containing 0.1% formic acid (gradient, total run-time 12.5 minutes). A representative total ion chromatogram of a human BALF extract is shown in Fig. 3A, and Fig. 3B and C show extracted ion chromatograms of C18:0-LPA and C18:0-LPS, respectively, demonstrating separation of the two species (as well as the significant contribution of endogenous C18:0-LPS to the C18:0-LPA channel). The small peak eluting before the most intense C18:0-LPS peak (sn1 isomer) in Fig. 3C represents the sn2 isomer; in general, separation between the sn2 and sn1 isomers was noted with all lysophospholipid species using the described chromatographic conditions. It is worth noting that the quality of separation between LPS and LPA, as well as the peak shape of LPA, varied among Kinetix C18 column batches. We also found that some new Kinetix columns required multiple injections of a biofluid extract in order to properly “condition” the column before adequate separation of LPS from LPA could be achieved, while other columns provided virtually baseline separation with no preconditioning. Further, column lifetime was limited regarding separation of LPS from LPA. Although the peak shape and separation of other lysophospholipid species remained excellent, the peak shape of LPA (and thus the separation from LPS) would begin to gradually deteriorate prior to other indicators of decreased column integrity (i.e., increased column pressure etc.). We also noted a tendency for LPA carry-over to increase with column life. Carry-over could be diminished with multiple injections of blank matrix (interestingly, solvent blanks did not contain LPA carry-over). Because new columns were always free of LPA, we believe that the carry-over was due to LPA build-up on the column (as opposed to build-up in the autosampler or other instrument tubing/valves). Column lifetime was variable, but in general, we found a need to change columns after approximately 500–700 sample injections. (Obviously, longer column life was preferable, but we were unable to identify a more robust chromatographic method that also effectively separated LPA from LPS.) Optimal separation was achieved at room temperature; column heating did narrow the peak width but also decreased chromatographic resolution. Effects of column cooling were not tested.
Fig. 3.
Total ion chromatogram (TIC) and extracted ion chromatograms (EIC) of a human BALF extract. Human BALF was extracted and analyzed by LC/MS/MS as described in Materials and Methods. The TIC is shown in A, the EIC of C18:0-LPA is shown in B, and the EIC for C18:0-LPS, showing separation of endogenous C18:0-LPA from signal contributed by endogenous C18:0-LPS, is shown in C.
Quantitation approach and method qualification using a stable isotope-labeled LPA standard as a surrogate calibrant
Quantitation of endogenous metabolites presents many challenges, including dealing with lack of a true matrix blank, how to generate relevant calibration curves in matrix containing an endogenous background, and, especially when a family of metabolites is of interest, lack of purchasable authentic standards. The LPAs of interest in clinical samples were C16:0-, C16:1-, C18:0-, C18:1-, C18:2-, C20:4-, and C22:6-LPA. Of these, authentic standards were available for C16:0-LPA, C18:0-LPA, C18:1-LPA, and C20:4-LPA only. For those LPAs for which no authentic standard was available, identification was based on accurate mass, MS/MS characteristics, chromatographic elution order, and retention time (relative to available LPA standards). To increase confidence in the peak assignment for C22:6-LPA, which tended to be of low abundance in biofluids and to suffer from close elution of multiple matrix peaks having the same MS/MS transition, we enzymatically synthesized C22:6-LPA as described under Materials and Methods and verified its retention time (data not shown).
Quantitation of LPA was based on use of an in-house synthesized stable isotope-labeled LPA (d5-C16:0-LPA) as a surrogate analyte for all LPAs of interest. The primary assumption of this approach was that the ionization efficiency of d5-C16:0-LPA was similar to that of other LPAs. We recognize the downside of this approach; if the ionization efficiencies among LPAs are not the same, errors will be introduced into the concentration values assigned. However, as our goal was to compare measured LPA concentration between two groups (and not to compare the concentration of individual LPA to each other), we felt that small bias in the reported concentrations was acceptable (given robust precision) and would not detract from achieving the desired goal of the analysis. Further, ionization comparisons of purchased LPA standards were made and there was no significant difference in ionization response noted (data not shown) although detailed investigations into the effect of different matrices and concentrations on ionization response were not conducted.
Calibration curves were generated by spiking d5-C16:0-LPA into human plasma or BALF at concentrations typically ranging from 0.24 (limit of quantitation, based on 0.2 ml of matrix) to 241 nM. The nonnaturally occurring LPA C17:0-LPA was used as an internal standard. “Qualification” (i.e., a fit-for-purpose partial validation based on, but not strictly adhering to, FDA bioanalytical method validation guidelines) of the method was performed based on the accuracy of quality controls (QC) generated by spiking plasma or BALF with known amounts of d5-C16:0-LPA (lower QC levels were chosen for BALF based on the observation that LPA levels in BALF were significantly less than in plasma). Stock solution stability (data not shown), intra-assay accuracy and precision, bench-top stability (conducted at one QC level only due to extensive bench-top evaluation described below), freeze-thaw stability, and autosampler stability of d5-C16:0-LPA in plasma and BALF were evaluated; the qualification results are summarized in Tables 1–3. Intra-day accuracy and precision, freeze-thaw stability, and autosampler stability for d5-C16:0-LPA in human plasma and BALF were acceptable (Tables 1 and 2). However, initial bench-top stability experiments were plagued with a consistent increase in the concentration of d5-C16:0-LPA over the length of the stability time-course (Table 3), which translated to low accuracy at time 0 but acceptable accuracy by 4 h. Investigations into the cause of this anomaly revealed that the method of IS addition was very important to achieving acceptable accuracy. In the initial sample preparation protocol, the IS was made in the LLE solvent (butanol) and added as a large volume to matrix. When the protocol was adjusted such that the IS was added as a small volume (i.e., 10 µl) to matrix, mixed, then subjected to butanol for extraction, the accuracy dramatically improved, suggesting that the method of introduction of an LPA into a biological matrix affects its distribution. Following adjustment of the IS addition protocol, the 4 h bench-top stability of d5-C16:0-LPA in human plasma and BALF met acceptance criteria (Tables 3 and 4).
TABLE 1.
d5-C16:0-LPA human plasma intra-day evaluation, freeze-thaw, and autosampler stability qualification results
| Intra-Day Evaluation | Freeze-Thaw Stability (3 cycles) | Autosampler Stability | ||||
| QC Level | Accuracy | Precision (% RSD) | Accuracy | Precision (% RSD) | Accuracy | Precision (% RSD) |
| QC1 | 97.7 | 7.9 | 94.7 | 1.7 | 98.7 | 7.2 |
| 72.2 nM | ||||||
| QC2 | 107 | 2.1 | 101 | 3.2 | 101 | 8.9 |
| 181 nM | ||||||
| QC3 | 92.3 | 9.1 | 97.3 | 3.2 | 87.2 | 4.5 |
| 481 nM | ||||||
TABLE 3.
d5-C16:0-LPA human plasma bench-top stability results
| [d5-C16:0-LPA] nM | Time (hours) | Accuracy: IS added in butanol extraction solvent (mean±SD) | Accuracy: IS (in MeOH) added as small volume (mean±SD) |
| 500 | 0 | 39.8 ± 6.1 | 107 ± 4.7 |
| 0.5 | 39.2 ± 8.6 | 105 ± 7.3 | |
| 1 | 58.9 ± 15.2 | 114 ± 8.4 | |
| 4 | 102 ± 20.4 | 116 ± 2.8 |
TABLE 2.
d5-C16:0-LPA human BALF intra-day evaluation, freeze-thaw, and autosampler stability qualification results
| Intra-Day Evaluation | Freeze-Thaw Stability (3 cycles) | Autosampler Stability | ||||
| QC Level | Accuracy | Precision (% RSD) | Accuracy | Precision (% RSD) | Accuracy | Precision (% RSD) |
| QC1 | 112 | 5.9 | 90.6 | 3.7 | 105 | 6.7 |
| 4.8 nM | ||||||
| QC2 | 95.6 | 2.6 | 89.6 | 4.7 | 109 | 4.6 |
| 36.1 nM | ||||||
| QC3 | 94.2 | 2.4 | 92.7 | 1.7 | 99.2 | 5.2 |
| 181 nM | ||||||
TABLE 4.
d5-C16:0-LPA human BALF bench-top stability results
| [d5-C16:0-LPA] nM | Time (hours) | Accuracy (Mean±SD) |
| 50 | 0 | 112 ± 9.5 |
| 0.5 | 105 ± 12.1 | |
| 1 | 116 ± 7.7 | |
| 4 | 92.3 ± 3.2 |
Evaluation of endogenous LPA bench-top and freeze-thaw stability
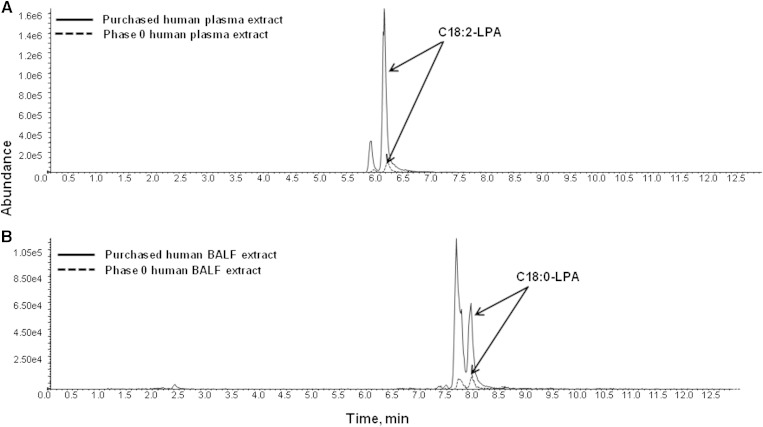
After successful qualification of d5-C16:0-LPA in human plasma and BALF, representative samples of plasma and BALF from the Phase 0 study were tested to verify that LPA levels in each biofluid were within range of the established calibration curve. Surprisingly, in both plasma and BALF, LPA levels were drastically lower than in multiple purchased lots of plasma and BALF used for method development and qualification (Fig. 4A, B). This led to investigation of the possibility that extensive and uncontrolled sample handling of purchased plasma and BALF caused LPA levels to increase through (presumably) enzymatic conversion. Even though d5-C16:0-LPA passed freeze-thaw and bench-top stability qualification, testing the stability of a stable isotope-labeled standard in biofluids only accounts for biological or chemical processes that lead to its degradation. Chemical or biological (i.e., enzymatic) pathways that lead to generation of the metabolites of interest (all of the measured LPAs) cannot be evaluated with stable isotope-labeled metabolites unless those pathways are well defined (which, in the case of LPA, they are not) and the appropriate stable isotope-labeled precursors can be added to the matrix. Therefore, to characterize the effects of sample handling on LPA stability, the freeze-thaw and bench-top stability of endogenous LPA was studied in plasma and BALF, comparing differences between fresh matrix and old or extensively handled matrix. “Old” refers to purchased plasma or BALF exposed to an unknown number of freeze-thaw cycles and lengths of time at room temperature. Fresh plasma refers to plasma obtained in-house from a volunteer pool and used on the day of collection (and kept on ice at all times). Because of the inherent difficulty in obtaining fresh BALF, we instead requested from the vendor (Bioreclamation) the freshest BALF possible and received BALF frozen immediately after collection. Therefore, baseline measurements from this BALF reflect exposure to one freeze/thaw cycle only (for simplicity we refer to this BALF as fresh).
Fig. 4.
EIC of C18:2-LPA (A) and C18:0-LPA (B) in purchased human BALF versus a Phase 0 BALF extract. Purchased human BALF or Phase 0 sample was extracted and analyzed by LC/MS/MS as described in Materials and Methods.
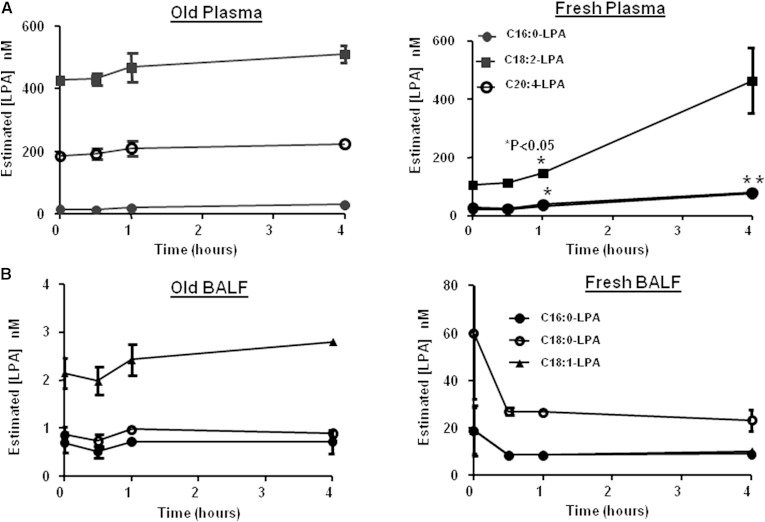
Table 5 shows the stability results from three representative endogenous LPAs in old human plasma and fresh human plasma subjected to freeze-thaw stability. In old plasma, the endogenous LPA levels did not significantly change over the course of three freeze-thaw cycles. In contrast, endogenous LPAs in fresh plasma showed a trend of increased levels over the course of the freeze-thaw cycles (C16:0-LPA and C18:2-LPA showed an initial decrease at the first freeze-thaw cycle vs. baseline) although the changes were not statistically significant except for C20:4-LPA at freeze-thaw 2 and 3. The increase varied among the different LPA species, with some species (C18:2-LPA and C20:4-LPA) doubling their concentration over the freeze-thaw period.
TABLE 5.
Freeze-thaw stability of endogenous LPA in fresh vs. old human plasma
| % Accuracy vs. Baseline Measurement mean±SD | ||||||
| LPA | Freeze-Thaw-1 | Freeze-Thaw-2 | Freeze-Thaw-3 | |||
| Old Plasma | Fresh Plasma | Old Plasma | Fresh Plasma | Old Plasma | Fresh Plasma | |
| C16:0-LPA | 99.6 ± 9.3 | 64.4 ± 14.4 | 90.4 ± 1.4 | 105 ± 19.1 | 94.9 ± 6.1 | 118 ± 21.9 |
| C18:2-LPA | 107.4 ± 5.7 | 65.3 ± 22.6 | 96.6 ± 1.5 | 97.3 ± 34.3 | 106.2 ± 4.2 | 129 ± 43.9 |
| C20:4-LPA | 107 ± 6.2 | 94.3 ± 17.3 | 96.9 ± 6.5 | 147 ± 23.6* | 105 ± 7.4 | 184 ± 35.5* |
*statistically significant from baseline measurement (one-way ANOVA, P < 0.05)
Table 6 shows the freeze-thaw stability results for three representative LPAs in old versus fresh human BALF. Similar to plasma, endogenous LPA in old BALF remained stable across three freeze-thaw cycles. In fresh BALF subjected to freeze-thaw, LPAs did not show the general trend of increased levels seen in fresh plasma, indicating that conversion of endogenous lysophospholipid to LPA may not be an issue in BALF. Rather, C16:0-LPA remained stable through three freeze-thaws. Although C18:0- and C18:1-LPA showed a decreased trend, variability in the baseline determination prevented statistical significance.
TABLE 6.
Freeze-thaw stability of endogenous LPA in fresh vs. old human BALF
| % Accuracy from Baseline Measurement mean±SD | ||||||
| LPA | Freeze-Thaw-1 | Freeze-Thaw-2 | Freeze-Thaw-3 | |||
| Old BALF | Fresh BALF | Old BALF | Fresh BALF | Old BALF | Fresh BALF | |
| C16:0-LPA | 117 ± 8.0 | 102 ± 19.9 | 105 ± 6.2 | 86.1 ± 4.4 | 91.0 ± 11.1 | 118 ± 44.2 |
| C18:0-LPA | 97.0 ± 9.3 | 53.7 ± 9.2 | 98.0 ± 7.9 | 50.9 ± 2.5 | 88.4 ± 6.8 | 64.2 ± 13.4 |
| C18:1-LPA | 109 ± 10.3 | 81.3 ± 5.9 | 111 ± 11.7 | 73.2 ± 2.6 | 99.8 ± 11.7 | 68.5 ± 2.6 |
Similar results were observed when old and fresh plasma or BALF was subjected to iced bench-top stability. Figure 5A shows the concentration profile of individual LPA in fresh versus old plasma over the course of 4 h. Comparable to the freeze-thaw stability results, an increase in LPA levels was evident in fresh plasma while levels remained stable in old plasma. In old BALF (Fig. 5B), LPA levels were stable over the stability time-course while in fresh BALF, the LPA showed a trend to initially decrease in the first 30 min (although the decrease was not statistically significant). Interestingly, fresh BALF heavily stressed (left at room temperature for at least 24 h then subjected to three freeze-thaw cycles) and then subjected to 4 h bench-top stability showed an LPA profile more similar to that seen in old BALF (data not shown).
Fig. 5.
Bench-top stability of endogenous LPA in human plasma/BALF. Purchased human plasma or BALF was subjected to bench-top stability, extracted, and analyzed by LC/MS/MS as described in Materials and Methods. A: stability of C16:0-, C18:2- and C20:4-LPA in old versus fresh plasma. B: stability of C16:0-, C18:0- and C18:1-LPA in old versus fresh BALF. P-values were calculated using a Student’s t-test (*P < 0.05).
Overall, our conclusion was that it is paramount to limit the freeze-thaw cycles and carefully plan the sample collection and analysis steps for future clinical and preclinical investigations.
Determination of LPA levels in BALF from a Phase 0 study
LPA levels were determined in plasma and BALF from a Phase 0 study comparing healthy controls (n = 6 plasma, n = 4 BALF) with IPF patients (n = 5 plasma, n = 4 BALF). In plasma (Fig. 6A), of the seven LPAs monitored, all were at levels above the LOQ except for C16:1-LPA. The most abundant LPA in Phase 0 human plasma was C18:2-LPA, consistent with results reported by other investigators (8, 17). The concentration of LPA ranged from approximately 0.7 to 13 nM. There was no difference in individual LPA levels between healthy controls and IPF patients.
Fig. 6.
Results from LPA analysis in Phase 0 plasma (A) and BALF (B). Phase 0 samples from healthy and IPF patients were extracted and analyzed by LC/MS/MS as described in Materials and Methods.
In BALF (Fig. 6B), C18:0-LPA was the only LPA present at levels above the LOQ (0.24 nM). C16:0-, C16:1-, C18:1-, and C18:2-LPA were detectable but below the LOQ and C20:4- and C22:6-LPA were not detectable. The concentration of C18:0-LPA ranged from approximately 153 to 594 pM, with a trend toward decreased levels in IPF patients.
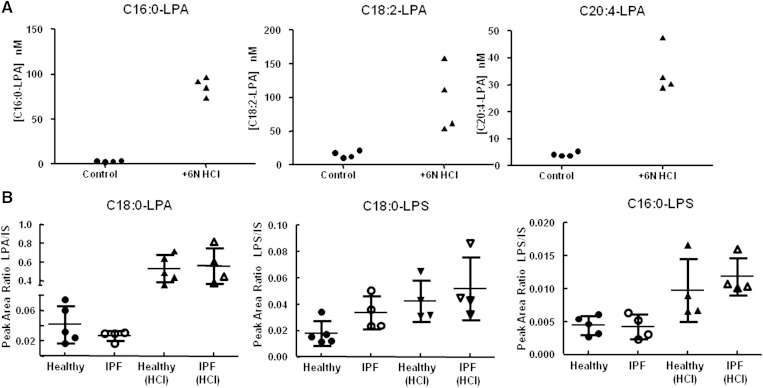
The concentration of LPA measured in Phase 0 plasma and BALF was noticeably lower than reported in the literature. For example, in human plasma, LPA levels determined using LC/MS techniques have typically been reported in the low to high µM range (8, 12–14, 28, 29) well above the LPA levels we measured. Though much less published data is available for LPA levels in human BALF, a direct comparison shows the LPA BALF concentrations we measured in Phase 0 samples to be significantly lower than what has been published to date (3, 18, 19). It should be noted that observed differences in BALF concentration were not entirely surprising because BALF composition is directly dependent on the sample processing procedures (i.e., bronchoscopy instillation volume), which can vary among institutions. However, one potential common denominator among the papers reporting LPA levels may be that sample preparation was conducted under highly acidic (typically 6N HCl) conditions, and/or LPAs were determined using nonselective methodology. Because acid has been shown to cause an increase in LPA formation due to head group hydrolysis from other phospholipids, we evaluated the effect of sample acidification on measured LPA levels in fresh human plasma and in leftover Phase 0 BALF. Plasma was extracted as described in Materials and Methods in the presence or absence of 6N HCl. For the LPA shown in Fig. 7A, the addition of 6N HCl resulted in a 29-, 6-, and 8-fold increase in LPA. BALF remaining from the Phase 0 samples was also extracted in the presence of 6N HCl. Figure 7B shows C18:0-LPA measured in Phase 0 healthy and IPF patient BALF extracted as described in Materials and Methods or in the presence of 6N HCl. The presence of HCl resulted in a 16-fold (healthy) and 21-fold (IPF) increase in C18:0-LPA. Because LPSs are the most likely lysophospholipids in BALF to interfere with LPA (based on our investigations), we also looked at the effect of acid on endogenous LPS species. Fig. 7B shows the effect of acid on C18:0-LPS and C16:0-LPS; acid caused a marginal (2- to 3-fold) increase in C18:0-LPS and C16:0-LPS.
Fig. 7.
Effect of acidification on LPA and LPS levels in purchased (A) and Phase 0 (B) BALF. Purchased BALF or Phase 0 BALF was extracted in the absence or presence of 6N HCl then analyzed by LC/MS/MS as described in Materials and Methods.
DISCUSSION
The observation that LPA levels in extensively handled plasma and BALF were higher than levels measured in the clinical samples was initially thought to be the result of endogenous enzymatic activity resulting in conversion of phospholipids or lysophospholipids to LPA. Extensive stability studies support that this could potentially be the case for plasma but not necessarily BALF. In human plasma, endogenous LPAs were shown to increase in freshly donated plasma subjected to bench-top or freeze-thaw stability studies but not in older plasma subjected to the same procedures (suggesting high enzymatic activity in fresh plasma and negligible enzymatic activity in old plasma). This increase in LPAs is consistent with observations made by other investigators and is assumed to be due to lysoPLD (i.e., autotaxin) activity (11, 13, 24–26). Further, in general, levels of endogenous LPAs were consistently higher in extensively handled plasma compared with fresh plasma. In contrast, no statistically significant differences in LPA levels were observed in newly purchased versus old BALF, suggesting that enzymatic activity might not be a contributing factor to LPA generation.
Beyond biological variation, we demonstrated that sample handling and sample preparation procedures can artificially increase measured levels of LPA. Concentrations of LPA in human plasma and BALF reported here were significantly lower than most LC/MS-based literature reports (low nM vs. high nM to µM). We believe this to be the result of the commonly followed procedure of including strong acid in lipid extraction protocols during sample preparation of biological fluids combined with use of partially-selective analytical methodology that does not adequately separate LPA from other interfering lipids. We showed that adding strong acid to human plasma or BALF during LLE can increase LPA levels as much as 30-fold. This increase was not simply a result of improved extraction efficiency as we verified that the extraction recovery of d5-C16:0-LPA and the IS were equivalent (∼60%) in the absence and presence of 6N HCl (data not shown). Moreover, in Phase 0 BALF acid was shown to increase not only LPA levels but also other endogenous lysophospholipids such as LPS, indicating that strong acid causes a cascade effect by which multiple lysophospholipid species may be formed from acid hydrolysis of lipid precursors. This cascade could further complicate interpretation of LPA changes in a patient population, especially as the lipomic profile (and therefore the pool of potential LPA precursors) likely varies among individuals. This further illustrates why resolution of LPA by either chromatography or selective isolation from other lipids during sample preparation is critical to ensure accurate measurement of LPA. Therefore, we would caution that analytical methods that employ acid and/or do not selectively separate LPA from other lipid species suffer from overestimation of LPA levels due to acid- or enzymatically-catalyzed conversion of lipids to LPA, and/or contributions to the LPA channel from coeluting lysophospholipid.
LPAs are important signaling molecules involved in a variety of cellular processes. Though many methods document LPA levels in plasma, reported levels of LPA in BALF are limited. Until more information is available, it is difficult to draw generalities around typical baseline levels of LPA or the relationship of LPA levels to a disease state or patient population. However, as more data become available, the methods used to determine LPA levels (as well as BALF collection procedures) should be considered carefully when comparing results between laboratories as the analytical methods and degree of biofluid dilution can dramatically affect the final reported concentration of LPA.
Acknowledgments
The authors acknowledge Sam Bonacorsi, Rich Burrell, and Wes Turley (Discovery Chemistry, Bristol-Myers Squibb, Princeton, NJ) for contributions to synthesis of stable isotope-labeled LPA standards and Jim Shen and Guowen Liu (Analytical and Bioanalytical Development, Bristol-Myers Squibb, Princeton, NJ) for many helpful discussions regarding testing and qualification of the LPA method. We are indebted to the Duke Immune Monitoring Core (Kelly Plonk, Alyssa Young, and Amber Beasock) and Duke University School of Medicine (Dr. Lake Morrison) for executing the Phase 0 clinical study and obtaining high-quality patient samples.
Footnotes
Abbreviations:
- BALF
- bronchoalveolar lavage fluid
- d5-C16:0-LPA
- d5-2-hydroxy-3-(phosphonooxy)propyl palmitate LPA
- IPF
- idiopathic pulmonary fibrosis
- IS
- internal standard
- LLE
- liquid-liquid extraction
- LOQ
- limit of quantitation
- LPA
- lysophosphatidic acid
- LPC
- lysophosphatidylcholine
- LPG
- lysophosphatidylglycerols
- LPI
- lysophosphatidylinositol
- LPS
- lysophosphatidylserine
- lysoPLD
- lysophospholipase D
REFERENCES
- 1.Gardell S. E., Dubin A. E., Chun J. 2006. Emerging medicinal roles for lysophospholipid signaling. Trends Mol. Med. 12: 65–75. [DOI] [PubMed] [Google Scholar]
- 2.Pradère J. P., Klein J., Gres S., Guigne C., Neau E., Valet P., Calise D., Chun J., Bascands J. L., Saulnier-Blache J. S., et al. 2007. LPA1 receptor activation promotes renal interstitial fibrosis. J. Am. Soc. Nephrol. 18: 3110–3118. [DOI] [PubMed] [Google Scholar]
- 3.Tager A. M., LaCamera P., Shea B. S., Campanella G. S., Selman M., Zhao Z., Polosukhin V., Wain J., Karimi-Shah B. A., Kim N. D., et al. 2008. The lysophosphatidic acid receptor LPA1 links pulmonary fibrosis to lung injury by mediating fibroblast recruitment and vascular leak. Nat. Med. 14: 45–54. [DOI] [PubMed] [Google Scholar]
- 4.Tigyi G. 2010. Aiming drug discovery at lysophosphatidic acid targets. Br. J. Pharmacol. 161: 241–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin M. E., Herr D. R., Chun J. 2010. Lysophosphatidic acid (LPA) receptors: signaling properties and disease relevance. Prostaglandins Other Lipid Mediat. 91: 130–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ley B., Collard H. R., King T. E., Jr 2011. Clinical course and prediction of survival in idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 183: 431–440. [DOI] [PubMed] [Google Scholar]
- 7.Jesionowska A., Cecerska E., Dolegowska B., B 2014. Methods for quantifying lysophosphatidic acid in biofluids: a review. Anal. Biochem. 453: 38–43. [DOI] [PubMed] [Google Scholar]
- 8.Smyth S. S., Cheng H. Y., Miriyala S., Panchatcharam M., Morris A. J. 2008. Roles of lysophosphatidic acid in cardiovascular physiology and disease. Biochim. Biophys. Acta. 1781: 563–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kishimoto T., Matsuoka T., Imamura S., Mizuno K. 2003. A novel colorimetric assay for the determination of lysophosphatidic acid in plasma using an enzymatic cycling method. Clin. Chim. Acta. 333: 59–67. [DOI] [PubMed] [Google Scholar]
- 10.Saulnier-Blache J. S., Girard A., Simon M. F., Lafontan M., Valet P. 2000. A simple and highly sensitive radioenzymatic assay for lysophosphatidic acid quantification. J. Lipid Res. 41: 1947–1951. [PMC free article] [PubMed] [Google Scholar]
- 11.Baker D. L., Desiderio D. M., Miller D. D., Tolley B., Tigyi G. J. 2001. Direct quantitative analysis of lysophosphatidic acid molecular species by stable isotope dilution electrospray ionization liquid chromatography-mass spectrometry. Anal. Biochem. 292: 287–295. [DOI] [PubMed] [Google Scholar]
- 12.Yoon H. R., Kim H., Cho S. H. 2003. Quantitative analysis of acyl-lysophosphatidic acid in plasma using negative ionization tandem mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 788: 85–92. [DOI] [PubMed] [Google Scholar]
- 13.Murph M., Tanaka T., Pang J., Felix E., Liu S., Trost R., Godwin A. K., Newman R., Mills G. 2007. Liquid chromatography mass spectrometry for quantifying plasma lysophospholipids: potential biomarkers for cancer diagnosis. Methods Enzymol. 433: 1–25. [DOI] [PubMed] [Google Scholar]
- 14.Shan L., Jaffe K., Li S., Davis L. 2008. Quantitative determination of lysophosphatidic acid by LC/ESI/MS/MS employing a reversed phase HPLC column. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 864: 22–28. [DOI] [PubMed] [Google Scholar]
- 15.Sutphen R., Xu Y., Wilbanks G. D., Fiorica J., Grendys E. C., Jr, LaPolla J. P., Arango H., Hoffman M. S., Martino M., Wakeley K., et al. 2004. Lysophospholipids are potential biomarkers of ovarian cancer. Cancer Epidemiol. Biomarkers Prev. 13: 1185–1191. [PubMed] [Google Scholar]
- 16.Liebisch G., Scherer M. 2012. Quantification of bioactive sphingo- and glycerophospholipid species by electrospray ionization tandem mass spectrometry in blood. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 883–884: 141–146. [DOI] [PubMed] [Google Scholar]
- 17.Baker D. L., Morrison P., Miller B., Riely C. A., Tolley B., Westermann A. M., Bonfrer J. M., Bais E., Moolenaar W. H., Tigyi G. 2002. Plasma lysophosphatidic acid concentration and ovarian cancer. JAMA. 287: 3081–3082. [DOI] [PubMed] [Google Scholar]
- 18.Georas S. N., Berdyshev E., Hubbard W., Gorshkova I. A., Usatyuk P. V., Saatian B., Myers A. C., Williams M. A., Xiao H. Q., Liu M., et al. 2007. Lysophosphatidic acid is detectable in human bronchoalveolar lavage fluids at baseline and increased after segmental allergen challenge. Clin. Exp. Allergy. 37: 311–322. [DOI] [PubMed] [Google Scholar]
- 19.Park G. Y., Lee Y. G., Berdyshev E., Nyenhuis S., Du J., Fu P., Gorshkova I. A., Li Y., Chung S., Karpurapu M., et al. 2013. Autotaxin production of lysophosphatidic acid mediates allergic asthmatic inflammation. Am. J. Respir. Crit. Care Med. 188: 928–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao Z., Xu Y. 2009. Measurement of endogenous lysophosphatidic acid by ESI-MS/MS in plasma samples requires pre-separation of lysophosphatidylcholine. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 877: 3739–3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tokumura A., Majima E., Kariya Y., Tominaga K., Kogure K., Yasuda K., Fukuzawa K. 2002. Identification of human plasma lysophospholipase D, a lysophosphatidic acid-producing enzyme, as autotaxin, a multifunctional phosphodiesterase. J. Biol. Chem. 277: 39436–39442. [DOI] [PubMed] [Google Scholar]
- 22.Aoki J., Inoue A., Okudaira S. 2008. Two pathways for lysophosphatidic acid production. Biochim. Biophys. Acta. 1781: 513–518. [DOI] [PubMed] [Google Scholar]
- 23.Okudaira S., Yukiura H., Aoki J. 2010. Biological roles of lysophosphatidic acid signaling through its production by autotaxin. Biochimie. 92: 698–706. [DOI] [PubMed] [Google Scholar]
- 24.Aoki J., Taira A., Takanezawa Y., Kishi Y., Hama K., Kishimoto T., Mizuno K., Saku K., Taguchi R., Arai H. 2002. Serum lysophosphatidic acid is produced through diverse phospholipase pathways. J. Biol. Chem. 277: 48737–48744. [DOI] [PubMed] [Google Scholar]
- 25.Nakamura K., Ohkawa R., Okubo S., Tozuka M., Okada M., Aoki S., Aoki J., Arai H., Ikeda H., Yatomi Y. 2007. Measurement of lysophospholipase D/autotaxin activity in human serum samples. Clin. Biochem. 40: 274–277. [DOI] [PubMed] [Google Scholar]
- 26.Scherer M., Schmitz G., Liebisch G. 2009. High-throughput analysis of sphingosine 1-phosphate, sphinganine 1-phosphate, and lysophosphatidic acid in plasma samples by liquid chromatography-tandem mass spectrometry. Clin. Chem. 55: 1218–1222. [DOI] [PubMed] [Google Scholar]
- 27.Zhao Y., Tong J., He D., Pendyala S., Evgeny B., Chun J., Sperling A. I., Natarajan V. 2009. Role of lysophosphatidic acid receptor LPA2 in the development of allergic airway inflammation in a murine model of asthma. Respir. Res. 10: 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim H., Yoon H-R., Pyo D. 2002. Quantitative analysis of lysophosphatic acid in human plasma by tandem mass spectrometry. Bull. Korean Chem. Soc. 23: 1139–1143. [Google Scholar]
- 29.Wang J., Sibrian-Vazquez M., Escobedo J. O., Lowry M., Wang L., Chu Y. H., Moore R. G., Strongin R. M. 2013. Simple enrichment and analysis of plasma lysophosphatidic acids. Analyst. 138: 6852–6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Das A. K., Milam J. E., Reddy R. C., Hajra A. K. 2006. Facile syntheses of acyl dihydroxyacetone phosphates and lysophosphatidic acids having different acyl groups. J. Lipid Res. 47: 1874–1880. [DOI] [PubMed] [Google Scholar]
- 31.Koivusalo M., Haimi P., Heikinheimo L., Kostiainen R., Somerharju P. 2001. Quantitative determination of phospholipid compositions by ESI-MS: effects of acyl chain length, unsaturation, and lipid concentration on instrument response. J. Lipid Res. 42: 663–672. [PubMed] [Google Scholar]
- 32.Montesi S. B., Mathai S. K., Brenner L. N., Gorshkova I. A., Berdyshev E. V., Tager A. M., Shea B. S. 2014. Docosatetraenoyl LPA is elevated in exhaled breath condensate in idiopathic pulmonary fibrosis. BMC Pulm. Med. 14: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zacarias A., Bolanowski D., Bhatnagar A. 2002. Comparative measurements of multicomponent phospholipid mixtures by electrospray mass spectroscopy: relating ion intensity to concentration. Anal. Biochem. 308: 152–159. [DOI] [PubMed] [Google Scholar]
- 34.Wishart D. S., Jewison T., Guo A. C., Wilson M., Knox C., Liu Y., Djoumbou Y., Mandal R., Aziat F., Dong E., et al. 2013. HMDB 3.0–The Human Metabolome Database in 2013. Nucleic Acids Res. 41: D801–D807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wishart D. S., Knox C., Guo A. C., Eisner R., Young N., Gautam B., Hau D. D., Psychogios N., Dong E., Bouatra S., et al. 2009. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res. 37: D603–D610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wishart D. S., Tzur D., Knox C., Eisner R., Guo A. C., Young N., Cheng D., Jewell K., Arndt D., Sawhney S., et al. 2007. HMDB: the Human Metabolome Database. Nucleic Acids Res. 35: D521–D526. [DOI] [PMC free article] [PubMed] [Google Scholar]