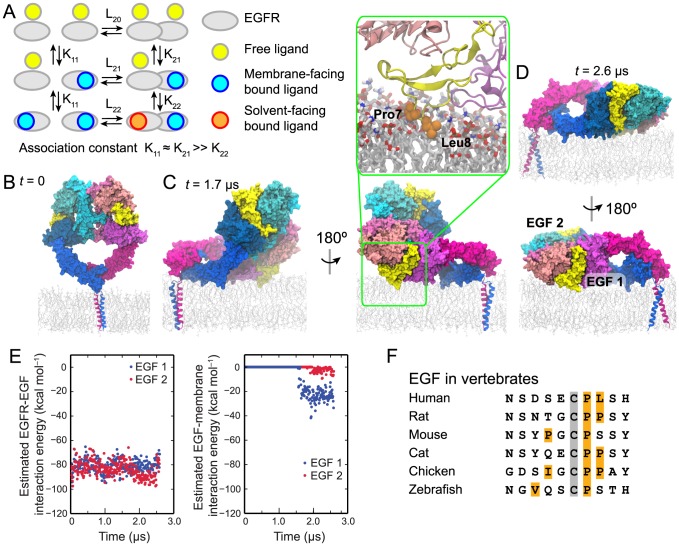
Figure 1. The ectodomain portion of the EGFR dimer lying on the membrane.
(A) A schematic description of EGFR ligand binding. The basic scheme is taken from ref. [15], but we have color coded the ligands to distinguish the free, the membrane-facing bound, and the solvent-facing bound ligands according to the simulation findings. “L” and “K” are used to denote the association constants of EGFR dimerization and ligand binding, respectively. The negative cooperativity is reflected in K21≫K22. The ligands bound to monomers are assumed to face the membrane as found in previous simulations of EGFR monomers (ref. [26]; see the Discussion). (B–D) Simulation of an EGFR dimer construct consisting of the ectodomains and TM helices. One EGFR subunit is colored in shades of blue, the other in shades of red, and the two bound EGF ligands in yellow. In the initial state (B), the ectodomain dimer is standing upright, perpendicular to the membrane. In the course of the simulation, the ectodomain dimer approaches the membrane (C), permitting the formation of extensive interactions between one of the ligands and the membrane, resulting in the partial-resting orientation. The EGF side chains Pro7 and Leu8 (orange) penetrate deep into the membrane, reaching the lipid tails (inset). Later in the course of the simulation (D), the ectodomain dimer approaches closer to the membrane and lies flat on its surface, in the full-resting orientation. (E) The free energy of each ligand's interaction with its host receptor (EGF1, blue; EGF2, red) in a two-ligand EGFR dimer (left panel) and of its interaction with the membrane bilayer (right panel); calculations used the MM/GBVI method. (F) The N-terminal sequences of EGF in various vertebrate species and from a set of members of the human EGF family. The amino acids shown correspond to residues 1–10 of human EGF. The conserved Cys6 is marked in gray and hydrophobic residues in positions 1–8 are marked in orange. Hydrophobic and aromatic residues beyond position 8 are expected to be buried in the protein interior.