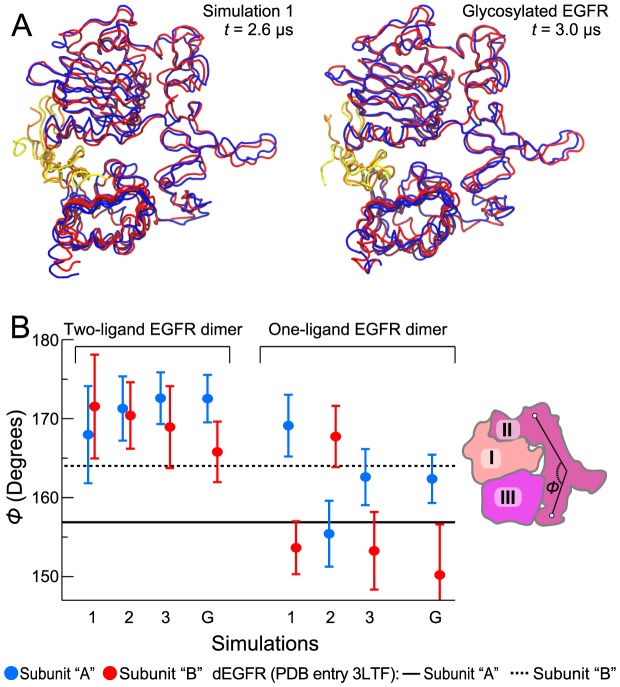
Figure 4. The two-ligand ectodomain dimer remaining symmetric in the simulations.
(A) The two subunits of the ectodomain dimer, as observed at the end of one of the simulations without glycosylation (left) and at the end of the simulation with full glycosylation (right), superimposed using the Cα atoms of domains I–III for reference. The EGFR ectodomains I–III are shown in blue and red, and the EGF molecules bound to them are shown in yellow and orange, respectively. (B) Domain II maintaining the same conformation in both subunits of the two-ligand EGFR dimer. Angle Φ (the angle formed by Cα atoms of residues 194, 239, and 296 in EGFR, or 189, 235, and 289 in dEGFR) characterizes the bending of domain II. This angle is different in each of the two subunits of the asymmetric two-ligand dEGFR dimer (the solid and dashed black lines; PDB entry 3LTF). The average angles in the simulations of two- and one-ligand EGFR dimers (Figs. 2, 3, and 5; labels refer to nonglycosylated EGFR simulations 1, 2, and 3, and glycosylated EGFR simulation G) are shown for each of the two subunits in blue and red (error bars correspond to the standard deviation). The angle Φ is illustrated in the schematic of domains I, II, and III of EGFR on the right.