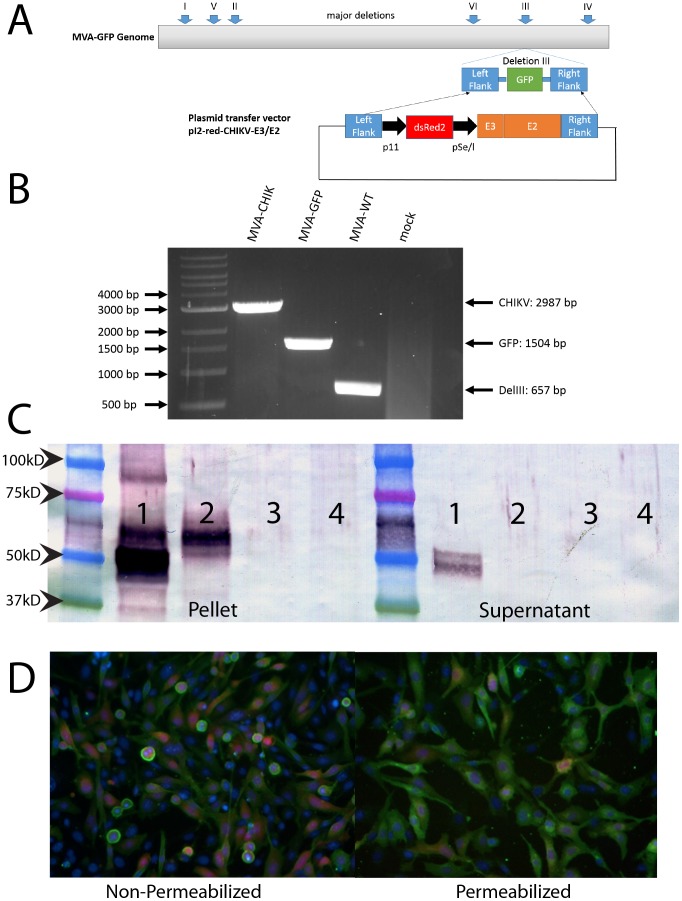
Figure 1. Generation and characterization of candidate MVA-CHIK vaccine virus.
Diagram of the MVA-GFP genome and transfer vector used for homologous recombination for creation of MVA-CHIK. The different areas where deletion occurred in the vaccinia virus genome to create MVA are shown. MVA-CHIK was created by transfecting the plasmid containing CHIKV p62 under control of a se/l promoter which also expressed dsRed2 under control of a p11 promoter. This plasmid contains flanking regions to deletion III (DelIII) which is where the recombinant gene was inserted into the genome with a fluorescent marker (A). PCR analysis of the DelIII region. Viral DNA was extracted from purified, final virus stocks of MVA-CHIK, MVA-GFP and MVA-WT. PCR was performed using primers specific for the DelIII flanking regions. A DNA ladder is included for comparison of size (B). Monolayers of CEF cells were infected with recombinant MVA-CHIK viruses at MOI of 5 or 10 PFU/cell for Western Blot or Immunostaining, respectively. After 24 h post infection, cells were harvested and subjected to SDS-PAGE followed by western blot analysis (C) or fixed with 2% PFA as described in the methods. Both the cellular pellet and supernatant were analyzed for expression of CHIKV E3/E2. The order is the same for both and is as follows, wild-type CHIKV (1), MVA-CHIK (2), MVA-GFP (3), and mock infected cells (4). Following fixation, cells for immunostaining were either allowed to remain intact or permeabilized with triton X-100 (D). Anti-CHIKV polyclonal rabbit was used for primary staining for both assays. Green represents CHIKV E2 positive cells, red is dsRed protein produced by the virus and blue is nuclear staining with Hoechst.