Abstract
In Drosophila, development of the compound eye is orchestrated by a network of highly conserved transcriptional regulators known as the retinal determination (RD) network. The retinal determination gene eyes absent (eya) is expressed in most cells within the developing eye field, from undifferentiated retinal progenitors to photoreceptor cells whose differentiation begins at the morphogenetic furrow (MF). Loss of eya expression leads to an early block in retinal development, making it impossible to study the role of eya expression during later steps of retinal differentiation. We have identified two new regulatory regions that control eya expression during retinal development. These two enhancers are necessary to maintain eya expression anterior to the MF (eya-IAM) and in photoreceptors (eya-PSE), respectively. We find that deleting these enhancers affects developmental events anterior to the MF as well as retinal differentiation posterior to the MF. In line with previous results, we find that reducing eya expression anterior to the MF affects several early steps during early retinal differentiation, including cell cycle arrest and expression of the proneural gene ato. Consistent with previous observations that suggest a role for eya in cell proliferation during early development we find that deletion of eya-IAM leads to a marked reduction in the size of the adult retinal field. On the other hand, deletion of eya-PSE leads to defects in cone and pigment cell development. In addition we find that eya expression is necessary to activate expression of the cone cell marker Cut and to regulate levels of the Hedgehog pathway effector Ci. In summary, our study uncovers novel aspects of eya-mediated regulation of eye development. The genetic tools generated in this study will allow for a detailed study of how the RD network regulates key steps in eye formation.
Introduction
The early Drosophila eye imaginal disc consists of pluripotent cells that are committed to a retinal fate and express high levels of eyeless (ey), a critical regulator of eye development. These pluripotent retinal progenitors begin differentiation along a moving boundary called the morphogenetic furrow (MF) at the posterior margin of the early third instar eye imaginal disc [1], [2]. Several key cellular events take place immediately anterior to the advancing MF, including exit from the cell cycle and arrest in the G1 phase [3]. At the same time, retinal progenitors anterior to the MF initiate expression of the proneural gene atonal (ato), which encodes a basic helix-loop-helix (bHLH) transcription factor (TF) necessary for the onset of photoreceptor differentiation [4]. Posterior to the MF, ato expression resolves into regularly spaced single cells, which differentiate into R8 photoreceptors, the founding cell of each ommatidium (unit eye). The remaining cell types of the adult eye are progressively recruited and differentiate around each founding R8 photoreceptor [5].
The RD network member eyes absent (eya) is a conserved transcription co-factor expressed in a broad band anterior to the MF and in all cells posterior to the MF [6] Mutations in eya such as eya1 and eya2 lead to eye specific loss of eya expression and gives rise to adult flies without eyes, suggesting that eya is necessary for eye development [7], [8]. eya1 and eya2 imaginal discs express ey suggesting that the retinal progenitors lacking eya expression are committed to the eye lineage [7]. However, cells lacking eya expression do not execute any further steps of eye development [7], [8], [9], including G1 arrest, MF initiation, or ato expression [10]. In clones of eya mutant tissue, retinal progenitors overproliferate and grow out of the plane of the eye disc; these cells eventually undergo apoptosis [10]. Due to the early blocks in retinal differentiation caused by loss of eya expression, the role eya may play in differentiating photoreceptors has been difficult to study. One way to bypass this early block and to identify the role of eya during later steps in retinal developmental is to generate single-celled eya mutant clones posterior to the MF. Although this approach has suggested a requirement for eya function in differentiating photoreceptors, it use is limited by the minimal clone size possible [10].
Previous studies suggest that Eya functions at least in part through forming a transcriptional regulatory complex with the homeodomain transcription factor Sine oculis (So), which directly activates expression of several regulators of retinal development [11], [12], [13], [14], [15]. Eya has been shown to physically interact with So [10], raising the possibility that Eya can regulate So targets during eye development. In line with this idea, forced expression of eya and so causes G1 arrest of proliferating retinal progenitors anterior to the MF [16]. Based on these observations it has been proposed that Eya plays an important role in several steps from initiation of the MF [9] and G1 arrest of retinal progenitors anterior to the MF [16], to terminal photoreceptor differentiation [10]. However, evidence for the role of eya during photoreceptor morphogenesis posterior to the MF is lacking. We have addressed this gap by the design and implementation of an eya genomic rescue construct that allows normal early (anterior) expression but blocks eya expression posterior to the MF, such that retinal differentiation is properly initiated but downstream developmental events are affected.
In this report we describe two new regulatory regions of eya, eya-IAM (Immediately Anterior to the MF) and eya-PSE (Photoreceptor Specific Expression). Reporter analysis of these regulatory regions using destabilized GFP (dGFP) suggests that eya-IAM is expressed in retinal progenitors immediately anterior to the MF, while eya-PSE is expressed in differentiating photoreceptors posterior to the MF in the third instar eye imaginal disc. We have analyzed the role of these elements in regulating eya expression during retinal development by deleting them in a previously characterized eya genomic rescue construct. As predicted, our analysis of eyaGRΔIAM and eyaGRΔPSE animals supports a role for eya during G1 arrest of retinal progenitors anterior to the MF as well as expression of the proneural gene ato. Our analysis also suggests a novel role for eya in the development of cone and pigment cells.
Materials and Methods
Construction of enhancer reporter lines
Enhancer fragments were amplified from a BAC carrying the eya genomic region. The enhancer fragments were then subcloned into a plasmid carrying destabilized GFP (dGFP) as a reporter and an attB site for site-specific integration [17]. The eya-IAM fragment was generated using IAM-F 5′ AGAGAATTCCAAACACCTGGCATTATCGCTTCATCTCGG and IAM-R 5′AGCGGATCCAGTTTCGTCTCCTCTTTTGCTGCCTCTTTG, the eya-PSE enhancer was generated using PSE-F 5'-ATTGAATTCGTCCAGAGTGGTGGTGGTGA and PSE R 5'-GATCTCGAGGTACGATTTGTGCGTGCG. These primers also indicate the limits of deletion. The resulting construct was integrated into the P{CaryP}attP2 site (abbreviated P2), which is located at 3L∶11,063,638, using φC31 integrase. Site-specific integration was confirmed by PCR with attB/attP primers [18].
Recombineering-induced deletions of enhancers in the eya genomic rescue
The eya genomic rescue (eyaGR) constructs carrying deletions of eya-IAM and eya-PSE enhancers were generated using recombineering as previously described [18]. The recombineered deletion transgenes were injected into P2, the same site used for the wild-type eyaGR [19], and site-specific integration was verified by PCR with attB/attP primers. We performed PCR on genomic DNA from the transgenic flies to verify the deletions.
Drosophila genotypes
All crosses were performed on standard cornmeal agar at 25°C. Heat shocks were performed at 37°C as described in Anderson et.al., 2012. To test the function of the eya enhancers in eye development, we crossed the mutant eyaGR constructs into the following mutant backgrounds: eya2 homozygotes, which specifically lack Eya expression in the eye disc [7], and eyacliIID/Df(2L)BSC354. eyacliIID is a null allele caused by a premature stop codon that causes recessive embryonic lethality [20], and Df(2L)BSC354 (hereafter referred to as Df(eya) removes the entire eya locus [21]. Consequently, eyacliIID/Df (eya) lacks all endogenous eya function. The eyaΔIAM and eyaΔPSE clones were generated by crossing eyacliIID, FRT 40/CyO; eyaGRΔIAM/TM6B,Tb1 or eyacliIID, FRT 40/CyO; eyaGRΔPSE/TM6B,Tb1 respectively with hsflp; P{w[+mC] = arm-LacZ}, FRT40/CyO animals. The RNAi stocks w; UAS-soRNAi (transformant ID104386) and w; UAS-eyaRNAi (transformant ID 108071) from VDRC were used [17], [22].
Immunohistochemistry of third instar eye discs, pupal eye discs, and imaging of adult eyes and plastic sections
Primary antibodies used were mouse anti-Eya (1∶200), anti-Cyclin B (1∶200), Anti-Ci (1∶200), anti-Cut(1∶100), rat anti-Elav (1∶500) (Developmental Studies Hybridoma Bank), guinea pig anti-Sens (1∶2000), guinea pig anti-Ato (1∶1000, gift from H. Bellen), Rabbit anti-β-Gal (1∶1000, Promega) and chick anti-GFP (1∶1000, Abcam). All secondary antibodies were made in goat and used at a final dilution of 1∶500; Cy3 and Cy5 labeled secondary antibodies were obtained from Jackson ImmunoResearch, the Alexa Fluor 488 (used at 1∶500) and Phalloidin Alexa Fluor 594 (used at 1∶500) was obtained from life technologies. Images were taken with a Zeiss LSM 510 confocal microscope and processed with ImageJ and Adobe Photoshop software. Immunohistochemistry on third instar eye discs and imaging of the adult eye were conducted as described previously [17]. Staining of 48 hr pupal eye discs and the plastic sections of adult eyes were generated as previously described [23].
Results
Identification of distinct regulatory elements controlling retinal expression of eya
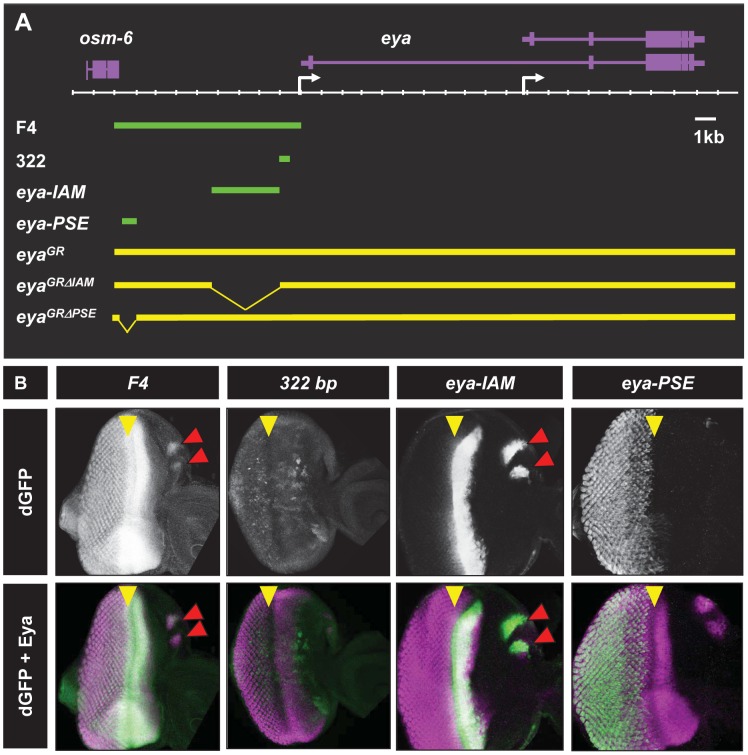
The eye-specific eya mutant eya2 harbors a 322 bp deletion approximately 500 bp upstream of the eya transcription start site [24]. This 322 bp deletion leads to a complete loss of eya expression in the eye imaginal disc, suggesting a necessary role for this region for eya expression during eye development. A previous report using LacZ reporter constructs suggests that the 322 bp region can recapitulate the basic pattern of eya expression anterior to the MF in the early larval eye disc [25]. However, the prolonged stability of LacZ and the low resolution of the activity staining method used in this study precluded a detailed understand of the transcriptional activity of the 322 bp segment. To further refine the transcriptional limits of this DNA, we tested the ability of this 322 bp region to drive destabilized GFP (dGFP) as a reporter. Due to its short half-life, dGFP has proven to be a far more accurate reporter of transcriptional activity than either eGFP of LacZ ([17], [26] and R.C. unpublished results). Consistent with previous reports, we find that the 322 bp enhancer is sufficient to drive early dGFP expression [25]. In contrast, however, our analysis suggests that 322 bp-driven dGFP expression in the early third instar is irregular (Figure S1A) by the early third instar stage, reporter expression begins to weaken (Figure S1C), and by mid to late third instar stage 322 bp driven dGFP expression is completely lost (Figure 1B). These results suggest that the 322 bp enhancer does not recapitulate the complete endogenous eya expression pattern during retinal development. Therefore, to identify the regulatory elements needed for complete retinal expression of eya we generated a large reporter construct, F4-dGFP that contains ∼9 kb of DNA upstream of the eya transcriptional start site (TSS), including the previously identified 322 bp enhancer (Figure 1A). We find that F4-dGFP recapitulates the complete endogenous eya expression pattern in the developing eye disc (Figure 1B and Figure S2A-A‴). In the second instar eye disc, F4-dGFP expression appears as a posterior to anterior gradient mimicking the endogenous eya gradient (data not shown). Posterior to the MF in third instar eye disc, F4 drives dGFP expression in differentiating photoreceptors and still maintains dGFP expression in retinal progenitors anterior to the MF (Figure 1B). Therefore, the ∼9 kb region upstream of the distal eya promoter harbors regulatory elements that are sufficient to recapitulate the endogenous eya expression in the developing eye imaginal disc.
Figure 1. Identification of two new cis retinal enhancers of eya.
A. Schematic of the eya locus showing enhancer fragments and genomic constructs. Genes (purple) are shown above the scale while the enhancer fragments (green bars) and genomic constructs (yellow) are on bottom. Transcription start sites are indicated by white arrows. B. Third instar eye discs showing enhancer-driven dGFP expression (white in top panels and green in bottom panels) along with endogenous Eya expression (magenta in bottom panels). Yellow arrowheads indicate the position of the MF and the red arrowheads mark ocellar dGFP expression in eya-IAM animals.
eya-IAM and eya-PSE enhancers are active during different stages of retinal development
The F4 region includes the 322 bp enhancer previously shown to be necessary for eya expression during eye development [24], [25]. To identify additional regulatory elements that are sufficient for complete retinal expression of eya, we analyzed different sub-fragments of F4. Through this analysis we have identified two separate regions that are named eya-IAM (Immediately Anterior to the MF) and eya-PSE (Photoreceptor Specific Enhancer), based on their ability to drive dGFP expression in undifferentiated and differentiating retinal cells, respectively (Figure 1A,B).
eya-IAM is a ∼3 kb fragment immediately upstream of the previously identified 322 bp enhancer (Figure 1A). eya-IAM expression begins as diffuse band at the posterior margin of the eye disc at ∼68 hr after egg laying (AEL, Figure S1B). After MF initiation, eya-IAM driven dGFP expression becomes more intense and is maintained at high levels anterior to the MF (Figure 1B). Both F4 as well as eya-IAM fragments drive the dGFP reporter expression that overlaps the broad band of endogenous eya expression immediately anterior to the MF (Figure 1B). These observations suggest that eya-IAM is sufficient for eya expression anterior to the MF.
The 785 bp eya-PSE fragment drives dGFP expression posterior to the MF in differentiating photoreceptors (Figure1B). eya-PSE driven dGFP expression is completely absent from second instar and early third larval instar eye discs (data not shown). eya-PSE driven dGFP co-localizes with Eya staining in photoreceptors posterior to the MF (Figure 1B). Co-staining with the neuronal differentiation marker Elav suggests that eya-PSE drives expression in the differentiating neurons (Figure 1B and Figure S3). eya-PSE driven dGFP expression is absent from undifferentiated cells posterior to the MF. Thus, eya-IAM and eya-PSE are sufficient to drive reporter gene expression closely mimicking the endogenous eya pattern, raising the possibility that they may also be required for proper eya expression during normal eye development.
eya-IAM is required for normal retinal development
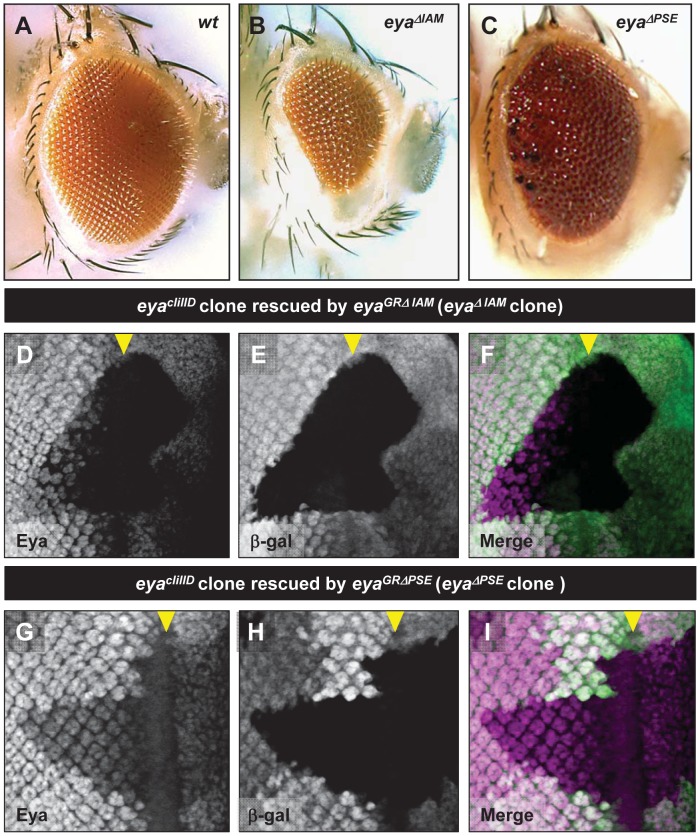
We tested if the eya-IAM enhancer is required for eya expression during normal eye development by deleting this region from a previously reported eya genomic rescue, eyaGR, that is sufficient to fully rescue all known eya mutant phenotypes (henceforth referred to as eyaGR, [19], [27]. The genomic rescue harboring the deletion of eya-IAM (eyaGRΔIAM, Figure 1A) rescues the lethality of eyacliIID/Df animals, however these rescued animals have a markedly reduced retinal field as compared to controls and wild type animals (Figure 2A, B and Figure S1H). Additionally, consistent with the ability of eya-IAM to drive expression of dGFP in ocelli (Figure 1B, red arrows) deletion of eya-IAM also leads to complete loss of ocelli in eyacliIID/Df animals (Figure S1D, E). The ability of eyaGRΔIAM to rescue the lethality of the eyacliIID allele suggests that eya-IAM may not play a major role in regulating the expression of eya during embryogenesis. Additionally, eyaGRΔIAM, partially rescues the complete loss of the retinal field observed in the eye specific eya allele eya2/Df(eya) animals (Figure S1J).
Figure 2. The IAM and PSE enhancers are required for eya expression during normal retinal development.
A-C. Adult eye phenotype of wild-type (A), eyacliIID/Df (eya); eyaGRΔIAM (B), eyacliIID/Df (eya); eyaGRΔPSE (rare escaper, C), D-I Clones of eyacliIID rescued by eyaGRΔIAM harboring a deletion of eya-IAM (D-F) and rescued by eyaGRΔPSE harboring a deletion of eya-PSE (G-I). The eyacliIID clones are marked by the absence β-galactosidase expression (white in E and H and green in I and J) showing reduced Eya expression (white in D and G and magenta in F and I). Yellow arrowheads indicate the position of the MF.
eya-IAM is required for normal eya expression anterior to the MF
Since eya-IAM is sufficient to drive strong reporter expression immediately anterior to the MF in mid-third instar eye discs, we tested if eya-IAM is required for normal Eya expression in vivo. This was analyzed in the eye discs of eyacliIID/Df; eyaGRΔIAM/+ animals (henceforth referred to as eyaΔIAM), as well as eyacliIID clones rescued by a single copy of eyaGRΔIAM (henceforth referred to as eyaΔIAM clones). As expected (assuming there are no other redundant enhancers), Eya expression immediately anterior to the MF in eyaΔIAM eye discs is significantly lower compared to control animals (Figure S1F,G). Moreover, eyaΔIAM clones show a complete loss of Eya expression anterior to the MF as compared to neighboring control tissue (Figure 2D-F). Both observations were verified in multiple eyaΔIAM eye discs and in eyaΔIAM anterior clones that do not span the MF (data not shown). Thus, consistent with the eya-IAM-dGFP reporter expression pattern, deletion of eya-IAM results in reduced eya expression in retinal progenitors anterior to the MF. In addition, eyaΔIAM clones posterior to the MF also show a reduction in the levels of Eya (Figure 2D-F), suggesting that in a genomic context eya-IAM may be required to maintain eya expression throughout retinal development. However the eyaΔIAM clones in the adult eye appear indistinguishable from heterozygous tissue. Therefore it is possible that loss of eya expression due to deletion of eya-IAM may not affect normal retinal development posterior to the MF. Consistent with this interpretation, sections of adult eyaΔIAM eyes show the normal complement of rhabdomeres (compare Figure 3C and D), suggesting that deletion of eya-IAM does not affect specification or differentiation of retinal cells posterior to the MF.
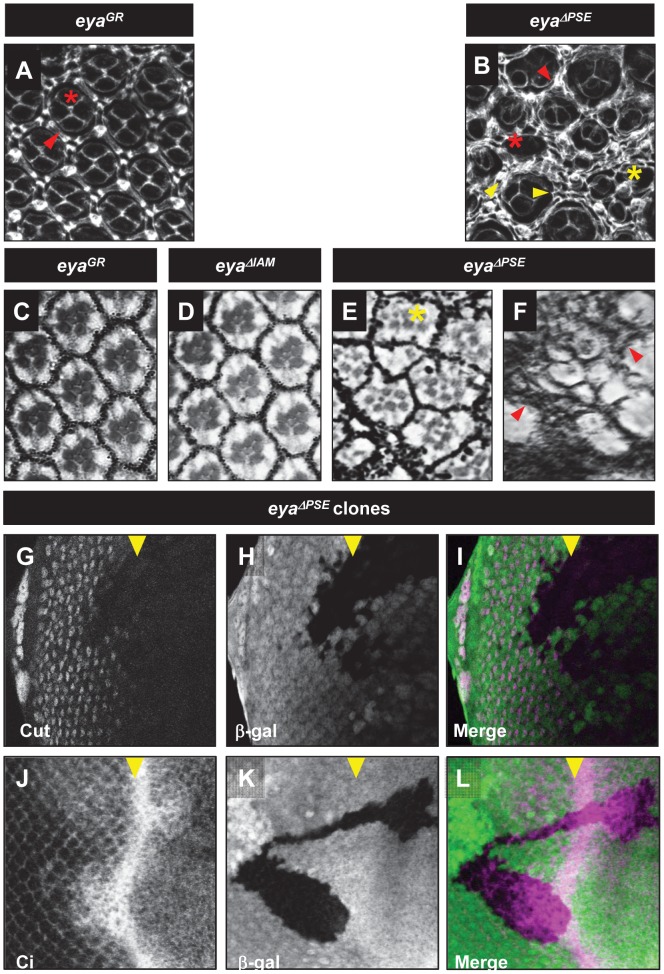
Figure 3. eya is required for development of cone and pigment cells.
Pupal retina (A, B), Adult plastic sections (C-F) of eyaGR and eyaΔPSE. Clones of eyaΔPSE in third instar eye disc (G-L). Pupae stained for Dlg (white, A and B) show cone cells and pigment cells. The adult eye sections show the rhabdomere structure of adult eyes. eyaGR (A and C) eyaGRΔPSE (B, E and F) eyaGRΔIAM (D). Arrow heads indicate pigment cells and asterisks show cone cells, respectively. Clones of eyacliIID rescued by eyaGRΔPSE stained for Cut (white in G, magenta in I) and Ci expression (white in J, magenta in L). eyacliIID clones are marked by the absence of β-galactosidase expression (white in H and K, green in I and L).
eya-PSE is required for normal retinal development
The eya-PSE enhancer is sufficient to drive dGFP expression in differentiating photoreceptors posterior to the MF. Therefore, we tested if eya-PSE is required for normal Eya expression posterior to the MF. We generated eyaGRΔ PSE, an eyaGR construct that harbors a deletion of eya-PSE (Figure 1A) and tested its ability to rescue retinal development in the absence of endogenous eya function. eyaGRΔPSE is largely unable to rescue the lethality of eyacliIID/Df animals, suggesting a role for the eya-PSE enhancer in additional tissues during development. However, rare eyacliIID/Df; eyaGRΔPSE/+ (henceforth referred to as eyaΔPSE) escapers do reach adulthood and eclose. Such eyaΔPSE escapers are unable to locomote normally and rapidly fall into the fly food and die. However, eyaΔPSE escapers do show strong eye defects, including a glazed appearance and occasional melanotic tissue on the eye surface. These defects are more severe toward the posterior margin of the eye as compared to the anterior region (Figure 2C). Consistent with this result, adult eyes of eyaΔPSE escapers show disorganized ommatidia in the anterior region (Figure 3E) while the posterior region shows a drastic loss of rhabdomeres and an excess of pigment cells (Figure 3F).
eya-PSE is required for normal retinal eya expression
To test if the eya-PSE deletion affects Eya expression posterior to the MF, we analyzed eyaΔPSE clones (eyacliIID clones rescued by a single copy of eyaGRΔPSE). In contrast to eyaΔPSE whole mutant escaper animals, eyaΔPSE clones show a clear reduction in the levels of Eya expression posterior to the MF (Figure 2G-I). eyaΔPSE clones that do not overlap the MF also show a severe reduction of eya expression (data not shown). As seen in Figure 2G-I, eyaΔPSE clones anterior to the MF also show a reduction in the levels of Eya expression, although not as severe as that seen in eyaΔIAMclones (compare Figure 2D and G). These observations have been verified in at least 5 independent clones for each genotype. The images in the figure 2D-I were captured and processed at identical settings to enable a direct comparison. Interestingly, while the eya-PSE enhancer is not sufficient to drive reporter expression in retinal cells anterior to the MF, eyaΔPSE clones anterior to the MF show a marked reduction in the levels of Eya (Figure 2D-F), suggesting that in the genomic context, eya-PSE may be required for normal Eya expression anterior to the MF. However, expression of Elav, a marker of photoreceptor differentiation, in eyaΔPSE clones is not significantly affected (Figure S2B) compared to eyaΔIAM clones (Figure 4G-I), suggesting that the reduction of eya expression anterior to the MF due to deletion of eya-PSE may not be sufficient to delay retinal specification and recruitment of photoreceptors.
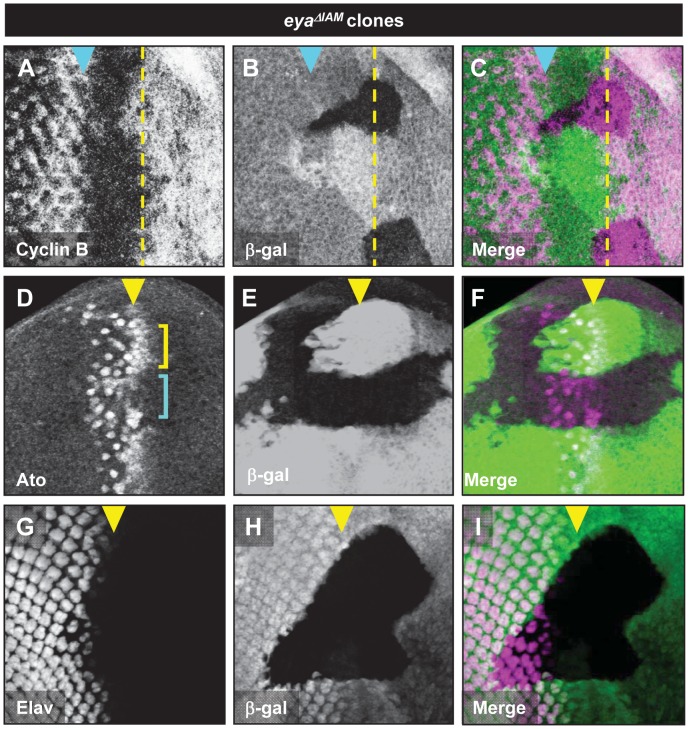
Figure 4. eya is required for normal Cyclin B and Ato expression.
Cyclin B (A), Ato (B) and Elav (C) expression in eyacliIID clones rescued by eyaGRΔIAM. eyacliIID clones are marked by the absence of β-galactosidase expression (green in B, C, E, F, H, and I). Yellow dashed lines and arrowheads indicate the position of the MF, cyan arrowheads in A-C indicate the position of the second mitotic wave. In panel D, cyan and yellow brackets mark Ato expression in eyacliIID clones and eyacliIID heterozygous tissue, respectively.
Normal Eya expression anterior to the MF is required for G1 arrest and initiation of retinal differentiation
Signals emanating from the MF play a critical role in the developmental events that take place anterior to the MF, such as mediating G1 arrest [28], [29], [30], [31] and induction of ato expression [29]. Loss of eya leads to a block of MF initiation and progression, precluding a detailed study of the role eya plays in the developmental events associated with MF movement. Since deletion of eya-IAM leads to a reduction of Eya and yet still allows initiation of retinal differentiation, we analyzed the effects of reducing eya expression anterior to the MF on both G1-arrest and ato induction. Specifically, we used eyaΔIAM clones to create retinal progenitors with reduced eya expression anterior to the MF.
G1 arrest was analyzed by monitoring Cyclin B expression. Normally, Cyclin B is absent from G1 arrested cells and is expressed in cells in the G2 and M phases [32]. We find that reduced levels of eya expression in eyaΔIAM clones leads to elevated Cyclin B expression in retinal progenitors that would normally undergo G1 arrest (Figure 4A-C). These results suggest that retinal progenitors with reduced levels of eya expression may not undergo efficient G1 arrest anterior to the MF. eyaΔIAM clones anterior to the MF also show decreased levels of ato expression in the pre-proneural zone (Figure 4D, compare the Ato expression indicated by cyan and the yellow brackets). While the defects in the Cyclin B and Ato expression due to reduced levels of eya are resolved as the MF progresses, these observations suggest that retinal progenitors with reduced levels of eya show a delay in developmental events prior to photoreceptor differentiation. Consistent with these observations, eyaΔIAM clones that span the MF also show a slight delay in the onset of photoreceptor differentiation (Figure 4G-I). Taken together, these observations suggest that eya expression is required for events associated with MF movement, both anterior and posterior to the MF.
eya expression is required for cone cell specification
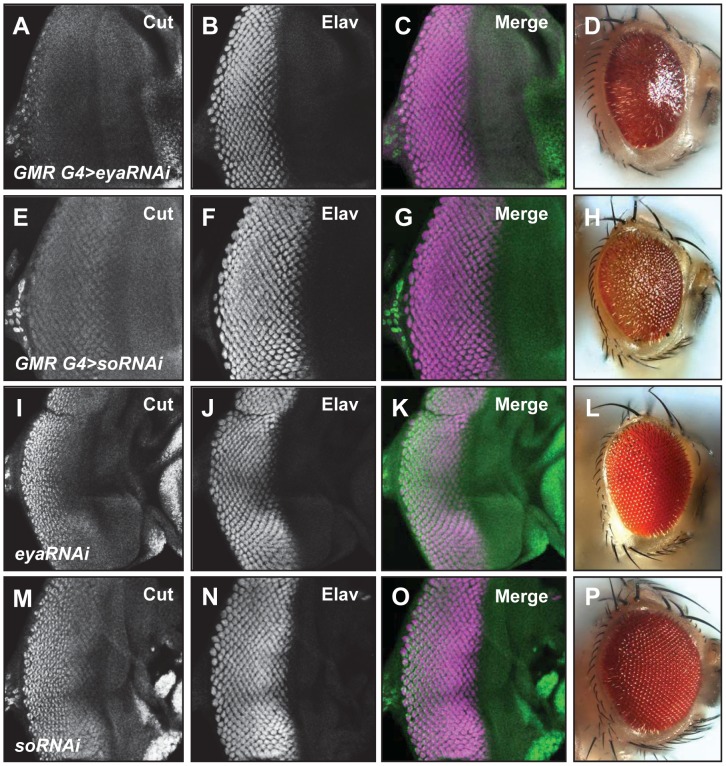
Very little is known about the role of eya in the development of support cells within ommatidia. Therefore, we used eyaΔPSE clones to test if eya is required for cone and pigment cell development. We analyzed cone cell specification in eyaΔPSE clones using Cut as an early marker for cone cell fate [33]. We find that eyaΔPSE clones do not express Cut (Figure 3G-I). We further tested the role of eya and its interaction partner so in regulating cone cell specification by knocking down the expression of eya and so using RNAi interference (RNAi) and analyzing Cut expression as marker for cone cell development. We used GMR-Gal4 to drive so-RNAi and eya-RNAi in all cells posterior to the MF. Knockdown of so or eya was confirmed by antibody staining (Figure S3A-D, E-H). We observe a severe reduction of Cut expression in animals with knockdown of either eya or so (Figure 5A, E), further supporting the hypothesis that eya may be required for cone cell development. Cut expression and eye development are normal in animals carrying the eya-RNAi or so-RNAi transgenes alone (Figure 5I-L, M-P). In addition, knockdown of eya or so does not significantly affect neuronal differentiation in the third instar eye disc as assayed by Elav staining (Figure 5B, F). However, adult eyes with so or eya knockdown have a glazed appearance, consistent with a cone cell defect (Figure 5D, 5H) [34]. To further test for a role for eya expression in cone cell development we examined pupal retinas of the rare eyaΔPSE animals. The pupal retinas of eyaΔPSE animals show a clear loss of cone cells (red asterisk in Figure 3B) compared to eyaGR animals (Figure 3A). Additionally, some ommatidia in the eyaΔPSE retina appeared to be fused (yellow asterisk in Figure 3B). Unexpectedly, pupal retinae of eyaΔPSE animals show an excess of interommatidial cells (compare arrowheads in Figure 3A and B). Consistent with these observations, sections of eyaΔPSE adult retinas show severely disorganized ommatidia with some ommatidia showing less than the normal complement of rhabdomeres as well as increased pigment cells (Figure 3E and F).
Figure 5. RNAi-mediated knockdown of eya or so strongly reduce Cut expression.
Cut (green) and Elav (magenta) expression in third instar eye discs (A-C, E-G, I-K, and M-O) and adult eyes (D, H, L and P) are shown. GMR-Gal4/+; UAS-eyaRNAi/+ (A-D), GMR-Gal4; UAS-soRNAi (E-H), UAS-eyaRNAi/+ (I-L), UAS-soRNAi/+ (M-P).
Loss of eya expression activates Hedgehog signaling posterior to the MF
Previous studies have suggested that ectopic expression of Hedgehog (Hh), as well as the Hh nuclear effector Cubitus interruptus (Ci), can cause an increase in the number of pigment cells in the Drosophila eye [35]. In addition, loss of Cul3 posterior to the MF causes elevated levels of Ci and also leads to an increase in the number of pigment cells [35]. Given the elevated numbers of pigment cells in eyaΔPSE animals, we investigated if elevated levels of Hh signaling persist in retinal cells harboring a deletion of eya-PSE. Hh is secreted by differentiating photoreceptors posterior to the MF and activates expression of target genes via the nuclear effector Cubitus interruptus (Ci) in the MF. Retinal progenitors in the MF stabilize the active form of Ci (CiAct) that activates transcription of Hh responsive genes such as dpp. However, Ci is actively degraded in all the cells posterior to the MF [36]. We investigated if reduced levels of eya expression would lead to higher levels of CiAct. We analyzed expression of Ci in eyaΔPSE clones using an antibody that recognizes both the full length and activated forms of Ci [37]. We find that reduced eya expression leads to elevated expression of Ci both anterior as well as posterior to the MF (Figure 3J-L), suggesting that eya may play a role in Ci degradation both anterior and posterior to the MF.
Regulation of eya-IAM and eya-PSE enhancers
eya receives regulatory input from several signaling pathways in the eye, as well as RD network members such as Ey [38], [39]. To identify potential direct regulators of eya-IAM and eya-PSE, we mutated putative binding sites of potential regulators within these enhancers, including Ey and Mad binding sites in eya-IAM, as well as So and several putative ETS binding sites in eya-PSE. Mad is the downstream effector of the Dpp pathway, which has been shown to activate Eya expression in the eye [9], and the ETS family transcription factor Pointed acts downstream of the Egfr pathway in differentiating eye cells [39]. None of these mutations caused any noticeable changes in enhancer activity (data not shown). Additionally, we tested if eya-IAM expression was altered in Mad1-2, smo3 double mutant clones that cannot respond to Dpp or Hh signaling in order to rule out indirect regulation by Dpp and Hh pathways. Consistent with the mutant binding site data, eya-IAM expression was unaltered in the Mad1-2, smo3 double mutant clones.
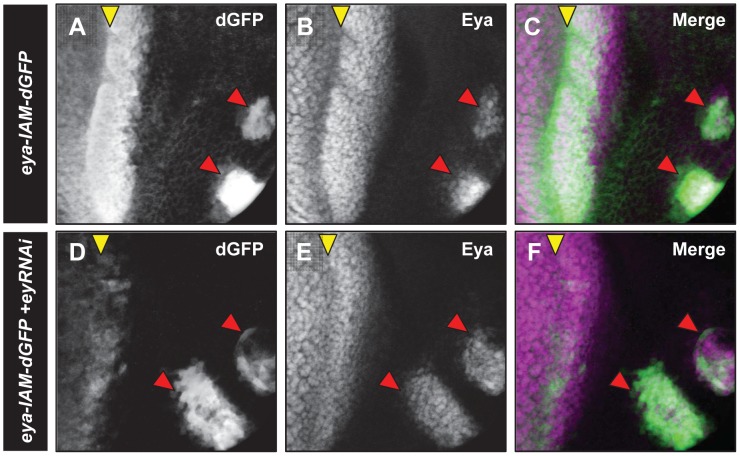
Mutation of a putative Ey binding site [40] did not alter eya-IAM driven dGFP expression. However previous reports show loss of Eya expression in ey mutant eye discs [41]. Therefore, it is possible that eya-IAM is regulated by Ey through a non-canonical site or indirectly via other transcription factors. To this end, we reduced levels of Ey anterior to the MF using ey-RNAi driven by hairyh10-Gal4 [42]. We find that in ey knockdown animals (ey-RNAi/eya-IAM; hairyh10-Gal4/+), dGFP expression anterior to the MF is markedly reduced (Figure 6). These results suggest that Ey regulates eya-IAM anterior to the MF, either directly or indirectly, through an unknown binding site.
Figure 6. eya-IAM-dGFP expression is regulated by ey immediately anterior to the MF.
Third instar eye discs showing dGFP expression (white in A and D, green in C and E) and endogenous Eya expression (white in B and E, magenta in C and E) in hairy-Gal4; eya-IAM-dGFP (A-C) and hairy-Gal4/ey-RNAi; eya-IAM-dGFP (D-F). Red arrowheads mark the position of developing ocelli.
Discussion
New tools to analyze the role of eya during retinal development
Many of the developmental events that take place anterior to the MF are dependent on each other for proper coordination and timing. For example, initiation of retinal specification marked by ato expression cannot take place without G1 arrest of retinal progenitors and G1 arrest in turn depends on the signaling from the MF. Loss of eya function in the developing eye disc blocks MF initiation and the remaining anterior retinal cells continue to proliferate and express ey during the early third instar stage [10], . Therefore, previously existing tools, such as clonal analysis using strong loss-of-function eya alleles, are not ideally suited to decipher the roles of eya in the plethora of developmental events that take place in retinal progenitors anterior to the MF or in differentiating cells posterior to the MF.
In this report we have identified two new cis enhancers of eya that are needed to regulate eya expression during eye development. Deletion of either the eya-IAM or eya-PSE enhancer leads to a reduction in the eye field or a glazed eye phenotype, respectively. eyaΔIAM and eyaΔPSE clones show significantly reduced levels of eya expression; together, these observations suggest that loss of these enhancers reduce eya expression while still allowing initiation of retinal differentiation. Thus, eyaΔIAM and eyaΔPSE clones have a distinct advantage over eyacliIID null clones in that they retain some ability to undergo retinal morphogenesis and differentiation. Therefore, eyaΔIAM and eyaΔPSE clones can be used to dissect developmental networks that are sensitive to the levels of Eya during retinal development, both anterior as well as posterior to the MF.
eya is required anterior to the MF to regulate G1 arrest and Ato expression
Gain- and loss-of-function studies have supported a role for eya in developmental processes anterior to the MF, including mediating G1 arrest of retinal progenitors [16]. The eyaΔIAM clones reported here represent a more refined system to study the role of eya in G1 arrest. Specifically, eyaΔIAM clones show reduced levels of eya but do not cause the complete block in retinal development that is associated with the eyacliIID null mutant clones. Our analysis suggests that reduced levels of Eya lead to delayed G1 arrest as evidenced by sustained Cyclin B expression in eyaΔIAM clones adjacent to the MF. This observation is consistent with previous studies that propose a role of eya in mediating the G1 arrest of retinal progenitors anterior to the MF [16].
Based on its physical interaction with the transcription factor So, Eya has been implicated in regulating ato expression. However, this role has never been directly tested.
Our analysis of the ato expression in eyaΔIAM clones shows a reduction of Ato in retinal progenitors with reduced eya expression. These results further support an important role for eya in mediating G1 arrest anterior to the MF and activating Ato expression during normal retinal development.
Cut regulation by Eya and So
Our results suggest that both eya and so are required for Cut expression in cone cells of the developing retina. This result differs from the recently published role of So-Eya in blocking Cut expression during specification of the antennal field in the second instar stage [43]. These seemingly opposite roles for So-Eya complex in Cut regulation are reminiscent of the recently reported ability of So to act both as a transcriptional activator of eyeless anterior to the MF as well as a transcriptional repressor of eyeless posterior to the MF [17]. Moreover, it is not surprising that the So-Eya complex may regulate certain genes in distinct ways during antennal specification versus cone cell development.
While the exact mechanism by which the So-Eya complex can activate or repress Cut expression is not known, our So ChIP-seq analysis does not show enrichment for So-binding at the cut locus [44]. However, lz has been shown to be a direct target of So and loss of lz leads to loss of Cut expression in the third instar disc posterior to the MF [14], [45]. Therefore, in the context of cone cell specification it is possible that the So-Eya complex may regulate Cut indirectly through direct regulation of lz.
Role of eya in downregulation of CiAct levels posterior to the MF
In absence of the Hh signal, Ci - the nuclear effector of the Hh pathway - is proteolytically cleaved to a transcriptional repressor form called CiRep. In the presence of the Hh signal this cleavage is blocked and leads to stabilization of the full length transcriptional activator form called CiAct. In the third instar eye disc, mature R8 photoreceptors posterior to the MF secrete Hh which diffuses to the MF and activates the Dpp signaling pathway. Thus, the full length form of Ci should be expressed in all cells posterior to the MF and within the MF. However, expression of CiAct is detected only in cells within the MF. It has been suggested that posterior to the MF, full length Ci is rapidly degraded by the Cul 3-Rdx complex [35], [36], [46]. Cul3 is a member of Drosophila Cullin family involved in protein degradation and Rdx is an adaptor protein encoded by roadkill (rdx). Loss of rdx function results in elevated expression of CiAct posterior to the MF [35], [36], [46], suggesting a critical role for rdx in the degradation of CiAct. An rdx-lacZ reporter suggests that rdx may be expressed in all cells posterior to the MF [47]; however, the molecular mechanism regulating rdx expression is not completely understood. It has been suggested that the EGFR pathway may play a role in regulation of rdx, although whether this activation is direct or indirect is not known [47]. Our results raise the possibility that eya expression maybe necessary for degradation of CiAct. Given that eya expression posterior to the MF is regulated by the EGFR pathway [39] and that rdx is expressed in the same cell that express eya, it is possible that Eya could mediate degradation of Ci via rdx activation. Consistent with this hypothesis, we found that rdx is enriched for So-binding (the binding partner of Eya) sites in third instar eye discs [44]. Thus, our observations, together with previous reports, raise the possibility that the So-Eya complex may be necessary for timely degradation of CiAct posterior to the MF. Previous work from our laboratory has shown that the proteolytically cleaved form of Ci – CiRep blocks activation of eya expression [48]. Thus a complex interplay between Eya and Hh may be required for precise regulation of eye development.
Mechanism for eya regulation of cone and pigment cell development
Our analysis of the rare eyaΔPSE escapers and the eyaΔPSE clones suggests a role for eya expression in cone and pigment cell development. Previous reports suggest that upregulation of Ci posterior to the MF can lead to excess pigment cells [35], [36], [46]. The upregulation of Ci in eyaΔPSE clones suggests that elevated Ci levels may contribute to excess pigment cells. However, reduced levels of eya posterior to the MF also cause defects in cone cell development. Signals from cone cells play an important role in specification of the primary pigment cell, thus raising the possibility that loss of cone cells may contribute to defective pigment cell development [3]. The tools generated in this study will be valuable in future studies of the mechanism by which eya regulates cone and pigment cell development.
In summary, we have identified two new cis regulatory regions of eya that are needed to maintain eya expression immediately anterior and posterior to the MF. We have shown that these enhancers are necessary and sufficient for proper regulation of eya expression during retinal development. Clonal analysis using eya genomic rescue constructs bearing deletions of these enhancers has shown for the first time that loss of eya results in a delay in G1 arrest of retinal progenitors anterior to the MF and a delay in the expression of the proneural protein ato. In addition, our results identify a potential role for eya in cone and pigment cell differentiation and timely degradation of Ci posterior to the MF.
Supporting Information
A-C. dGFP expression in larval eye discs driven by the 322 bp enhancer at 60 hr and 74 hr AEL (A and C) and the eya-IAM enhancer in a 60 hr eye disc (B). Eye discs were stained for dGFP (white in A and B, green in C) and endogenous Eya (red in C). D-E Ocelli of eyacliIID/CyO; eyaGRΔIAM (C) and eyacliIID/Df(eya); eyaGRΔIAM (D) animals. Yellow arrowheads indicate the positions of ocelli. F-G Eya expression in third instar eye discs (white in E and F) in eya2/CyO; eyaGRΔIAM (E), and eya2/Df(eya); eyaGRΔIAM (F). H-J. Adult eye phenotypes of eyacliIID/CyO; eyaGRΔIAM (H), eyacliIID/CyO; eyaGRΔPSE (I) and eya2/Df(eya); eyaGRΔIAM (J) animals.
(TIF)
F4-dGFP completely recapitulates the endogenous Eya expression pattern in mid-late third instar eye imaginal discs (A-A′″). dGFP expression (white in A and green in A''') completely overlaps with the endogenous Eya expression (white in A′ and magenta in A''') and the neuronal differentiation marker Elav (white in A″ and red in A'''). Overlap of all three markers is shown in A'''. The F4-dGFP eye disc shown here is the same as the one shown in Figure 1B. Elav (white in B″) expression in eyaΔPSE clone is not significantly altered. Eya expression is reduced (white in B′). Merge is shown in B′″. The clones shown here are the same as those shown in Figure 2G-I. C-F. The eya-PSE enhancer is active in differentiating photoreceptors posterior to the MF. Top panels (C-F) show third instar eye discs from transgenic larva carrying eya-PSE-dGFP. Eye discs were stained for dGFP (C and C′), Elav (D and D′), and Actin (E and E′). The bottom panels show orthogonal views of the same disc. The dotted yellow line in panel D indicates the position of the orthogonal sections.
(TIF)
A-H. RNAi knockdown of so and eya. Third instar eye discs of soRNAi/GMR-Gal4 (A-D) and eya-RNAi//GMR-Gal4 (E-H) stained for So (A and F), Eya (B and E), and Elav (C and G). Although So and Eya expression are clearly reduced in response to their respective RNAi constructs, no obvious change in ELAV expression is observed in either case.
(TIF)
Acknowledgments
UCK would like to thank past members of Mardon lab (2006–2012) for their kind support and help. We are grateful for technical help provided by Xuan Zhu and Meredith Bunkers. In addition we would like to thank members of the Chen lab for their support.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by the Retina Research Foundation (http://retinaresearchfnd.org/), the National Eye Institute (NEI) (www.nei.nih.gov) grant R01 EY011232 and the NEI/National Institutes of Health Core Grant for Vision Research EY-002520. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Cagan RL, Ready DF (1989) The emergence of order in the Drosophila pupal retina. Dev Biol 136: 346–362. [DOI] [PubMed] [Google Scholar]
- 2. Frankfort BJ, Mardon G (2002) R8 development in the Drosophila eye: a paradigm for neural selection and differentiation. Development 129: 1295–1306. [DOI] [PubMed] [Google Scholar]
- 3. Kumar JP (2012) Building an ommatidium one cell at a time. Dev Dyn 241: 136–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jarman AP, Grell EH, Ackerman L, Jan LY, Jan YN (1994) Atonal is the proneural gene for Drosophila photoreceptors. Nature 369: 398–400. [DOI] [PubMed] [Google Scholar]
- 5. Yang L, Baker NE (2001) Role of the EGFR/Ras/Raf pathway in specification of photoreceptor cells in the Drosophila retina. Development 128: 1183–1191. [DOI] [PubMed] [Google Scholar]
- 6. Bonini NM, Leiserson WM, Benzer S (1998) Multiple roles of the eyes absent gene in Drosophila. Dev Biol 196: 42–57. [DOI] [PubMed] [Google Scholar]
- 7. Bonini NM, Leiserson WM, Benzer S (1993) The eyes absent gene: genetic control of cell survival and differentiation in the developing Drosophila eye. Cell 72: 379–395. [DOI] [PubMed] [Google Scholar]
- 8. Leiserson WM, Benzer S, Bonini NM (1998) Dual functions of the Drosophila eyes absent gene in the eye and embryo. Mech Dev 73: 193–202. [DOI] [PubMed] [Google Scholar]
- 9. Curtiss J, Mlodzik M (2000) Morphogenetic furrow initiation and progression during eye development in Drosophila: the roles of decapentaplegic, hedgehog and eyes absent. Development 127: 1325–1336. [DOI] [PubMed] [Google Scholar]
- 10. Pignoni F, Hu B, Zavitz KH, Xiao J, Garrity PA, et al. (1997) The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development. Cell 91: 881–891. [DOI] [PubMed] [Google Scholar]
- 11. Hayashi T, Xu C, Carthew RW (2008) Cell-type-specific transcription of prospero is controlled by combinatorial signaling in the Drosophila eye. Development 135: 2787–2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pappu KS, Ostrin EJ, Middlebrooks BW, Sili BT, Chen R, et al. (2005) Dual regulation and redundant function of two eye-specific enhancers of the Drosophila retinal determination gene dachshund. Development 132: 2895–2905. [DOI] [PubMed] [Google Scholar]
- 13. Pauli T, Seimiya M, Blanco J, Gehring WJ (2005) Identification of functional sine oculis motifs in the autoregulatory element of its own gene, in the eyeless enhancer and in the signalling gene hedgehog. Development 132: 2771–2782. [DOI] [PubMed] [Google Scholar]
- 14. Yan H, Canon J, Banerjee U (2003) A transcriptional chain linking eye specification to terminal determination of cone cells in the Drosophila eye. Dev Biol 263: 323–329. [DOI] [PubMed] [Google Scholar]
- 15. Zhang T, Ranade S, Cai CQ, Clouser C, Pignoni F (2006) Direct control of neurogenesis by selector factors in the fly eye: regulation of atonal by Ey and So. Development 133: 4881–4889. [DOI] [PubMed] [Google Scholar]
- 16. Bessa J, Gebelein B, Pichaud F, Casares F, Mann RS (2002) Combinatorial control of Drosophila eye development by eyeless, homothorax, and teashirt. Genes Dev 16: 2415–2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Atkins M, Jiang Y, Sansores-Garcia L, Jusiak B, Halder G, et al. (2013) Dynamic rewiring of the Drosophila retinal determination network switches its function from selector to differentiation. PLoS Genet 9: e1003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Venken KJ, He Y, Hoskins RA, Bellen HJ (2006) P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314: 1747–1751. [DOI] [PubMed] [Google Scholar]
- 19. Jin M, Jusiak B, Bai Z, Mardon G (2013) Eyes absent tyrosine phosphatase activity is not required for Drosophila development or survival. PLoS One 8: e58818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nusslein-Volhard C, Wieschaus E (1980) Mutations affecting segment number and polarity in Drosophila. Nature 287: 795–801. [DOI] [PubMed] [Google Scholar]
- 21. Cook RK, Christensen SJ, Deal JA, Coburn RA, Deal ME, et al. (2012) The generation of chromosomal deletions to provide extensive coverage and subdivision of the Drosophila melanogaster genome. Genome Biol 13: R21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, et al. (2007) A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448: 151–156. [DOI] [PubMed] [Google Scholar]
- 23. Pepple KL, Anderson AE, Frankfort BJ, Mardon G (2007) A genetic screen in Drosophila for genes interacting with senseless during neuronal development identifies the importin moleskin. Genetics 175: 125–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zimmerman JE, Bui QT, Liu H, Bonini NM (2000) Molecular genetic analysis of Drosophila eyes absent mutants reveals an eye enhancer element. Genetics 154: 237–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bui QT, Zimmerman JE, Liu H, Gray-Board GL, Bonini NM (2000) Functional analysis of an eye enhancer of the Drosophila eyes absent gene: differential regulation by eye specification genes. Dev Biol 221: 355–364. [DOI] [PubMed] [Google Scholar]
- 26. Li Y, Jiang Y, Chen Y, Karandikar U, Hoffman K, et al. (2013) optix functions as a link between the retinal determination network and the dpp pathway to control morphogenetic furrow progression in Drosophila. Dev Biol 381: 50–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jusiak B, Abulimiti A, Haelterman N, Chen R, Mardon G (2012) MAPK target sites of eyes absent are not required for eye development or survival in Drosophila. PLoS One 7: e50776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Baonza A, Freeman M (2005) Control of cell proliferation in the Drosophila eye by Notch signaling. Dev Cell 8: 529–539. [DOI] [PubMed] [Google Scholar]
- 29. Dominguez M (1999) Dual role for Hedgehog in the regulation of the proneural gene atonal during ommatidia development. Development 126: 2345–2353. [DOI] [PubMed] [Google Scholar]
- 30. Escudero LM, Freeman M (2007) Mechanism of G1 arrest in the Drosophila eye imaginal disc. BMC Dev Biol 7: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Firth LC, Baker NE (2005) Extracellular signals responsible for spatially regulated proliferation in the differentiating Drosophila eye. Dev Cell 8: 541–551. [DOI] [PubMed] [Google Scholar]
- 32. Lopes CS, Casares F (2010) hth maintains the pool of eye progenitors and its downregulation by Dpp and Hh couples retinal fate acquisition with cell cycle exit. Dev Biol 339: 78–88. [DOI] [PubMed] [Google Scholar]
- 33. Blochlinger K, Jan LY, Jan YN (1993) Postembryonic patterns of expression of cut, a locus regulating sensory organ identity in Drosophila. Development 117: 441–450. [DOI] [PubMed] [Google Scholar]
- 34. Pickup AT, Ming L, Lipshitz HD (2009) Hindsight modulates Delta expression during Drosophila cone cell induction. Development 136: 975–982. [DOI] [PubMed] [Google Scholar]
- 35. Ou CY, Wang CH, Jiang J, Chien CT (2007) Suppression of Hedgehog signaling by Cul3 ligases in proliferation control of retinal precursors. Dev Biol 308: 106–119. [DOI] [PubMed] [Google Scholar]
- 36. Mistry H, Wilson BA, Roberts IJ, O'Kane CJ, Skeath JB (2004) Cullin-3 regulates pattern formation, external sensory organ development and cell survival during Drosophila development. Mech Dev 121: 1495–1507. [DOI] [PubMed] [Google Scholar]
- 37. Motzny CK, Holmgren R (1995) The Drosophila cubitus interruptus protein and its role in the wingless and hedgehog signal transduction pathways. Mech Dev 52: 137–150. [DOI] [PubMed] [Google Scholar]
- 38. Firth LC, Baker NE (2009) Retinal determination genes as targets and possible effectors of extracellular signals. Dev Biol 327: 366–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Salzer CL, Kumar JP (2010) Identification of retinal transformation hot spots in developing Drosophila epithelia. PLoS One 5: e8510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ostrin EJ, Li Y, Hoffman K, Liu J, Wang K, et al. (2006) Genome-wide identification of direct targets of the Drosophila retinal determination protein Eyeless. Genome Res 16: 466–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Halder G, Callaerts P, Flister S, Walldorf U, Kloter U, et al. (1998) Eyeless initiates the expression of both sine oculis and eyes absent during Drosophila compound eye development. Development 125: 2181–2191. [DOI] [PubMed] [Google Scholar]
- 42. Ellis MC, Weber U, Wiersdorff V, Mlodzik M (1994) Confrontation of scabrous expressing and non-expressing cells is essential for normal ommatidial spacing in the Drosophila eye. Development 120: 1959–1969. [DOI] [PubMed] [Google Scholar]
- 43. Wang CW, Sun YH (2012) Segregation of eye and antenna fates maintained by mutual antagonism in Drosophila. Development 139: 3413–3421. [DOI] [PubMed] [Google Scholar]
- 44. Jusiak B, Karandikar UC, Kwak SJ, Wang F, Wang H, et al. (2014) Regulation of Drosophila Eye Development by the Transcription Factor Sine oculis. PLoS One 9: e89695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Flores GV, Duan H, Yan H, Nagaraj R, Fu W, et al. (2000) Combinatorial signaling in the specification of unique cell fates. Cell 103: 75–85. [DOI] [PubMed] [Google Scholar]
- 46. Kent D, Bush EW, Hooper JE (2006) Roadkill attenuates Hedgehog responses through degradation of Cubitus interruptus. Development 133: 2001–2010. [DOI] [PubMed] [Google Scholar]
- 47. Baker NE, Bhattacharya A, Firth LC (2009) Regulation of Hh signal transduction as Drosophila eye differentiation progresses. Dev Biol 335: 356–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pappu KS, Chen R, Middlebrooks BW, Woo C, Heberlein U, et al. (2003) Mechanism of hedgehog signaling during Drosophila eye development. Development 130: 3053–3062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A-C. dGFP expression in larval eye discs driven by the 322 bp enhancer at 60 hr and 74 hr AEL (A and C) and the eya-IAM enhancer in a 60 hr eye disc (B). Eye discs were stained for dGFP (white in A and B, green in C) and endogenous Eya (red in C). D-E Ocelli of eyacliIID/CyO; eyaGRΔIAM (C) and eyacliIID/Df(eya); eyaGRΔIAM (D) animals. Yellow arrowheads indicate the positions of ocelli. F-G Eya expression in third instar eye discs (white in E and F) in eya2/CyO; eyaGRΔIAM (E), and eya2/Df(eya); eyaGRΔIAM (F). H-J. Adult eye phenotypes of eyacliIID/CyO; eyaGRΔIAM (H), eyacliIID/CyO; eyaGRΔPSE (I) and eya2/Df(eya); eyaGRΔIAM (J) animals.
(TIF)
F4-dGFP completely recapitulates the endogenous Eya expression pattern in mid-late third instar eye imaginal discs (A-A′″). dGFP expression (white in A and green in A''') completely overlaps with the endogenous Eya expression (white in A′ and magenta in A''') and the neuronal differentiation marker Elav (white in A″ and red in A'''). Overlap of all three markers is shown in A'''. The F4-dGFP eye disc shown here is the same as the one shown in Figure 1B. Elav (white in B″) expression in eyaΔPSE clone is not significantly altered. Eya expression is reduced (white in B′). Merge is shown in B′″. The clones shown here are the same as those shown in Figure 2G-I. C-F. The eya-PSE enhancer is active in differentiating photoreceptors posterior to the MF. Top panels (C-F) show third instar eye discs from transgenic larva carrying eya-PSE-dGFP. Eye discs were stained for dGFP (C and C′), Elav (D and D′), and Actin (E and E′). The bottom panels show orthogonal views of the same disc. The dotted yellow line in panel D indicates the position of the orthogonal sections.
(TIF)
A-H. RNAi knockdown of so and eya. Third instar eye discs of soRNAi/GMR-Gal4 (A-D) and eya-RNAi//GMR-Gal4 (E-H) stained for So (A and F), Eya (B and E), and Elav (C and G). Although So and Eya expression are clearly reduced in response to their respective RNAi constructs, no obvious change in ELAV expression is observed in either case.
(TIF)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.