Abstract
In this work, we present a simple and fast approach for simultaneous detection of nucleic acid and protein using gold nanoparticles (GNPs) and a lateral flow device (LFD). Sandwich-type immunoreactions and DNA hybridizations were performed simultaneously on the LFD by using DNA- and antibody-functionalized GNPs. The captured GNPs, due to the DNA hybridization and immunoreaction events on the LFD, produced characteristic red bands that could be used for the qualitative detections of DNA and/or protein. The proof of principle was demonstrated by using 60-mer DNA and rabbit IgG (R-IgG) model targets. The LFD was capable of detecting a minimum of 0.5 nM target DNA and 2 ng mL−1 IgG simultaneously in 15 min. The proposed LFD shows great promise for in-field and point-of-care testing of disease-related circulating nucleic acid and protein biomarkers in biological fluids.
Keywords: Lateral flow, nucleic acid, protein, gold nanoparticle
Introduction
Noninvasive detection of serum biomarkers including circulating proteins and nucleic acids in biological fluids is of central importance for early disease diagnosis, screening and staging of diseases, as well as the evaluation of response to therapy.1–3 Serum protein biomarker screening lacks sensitivity and specificity, and their role in early diagnosis and treatment remains to be elucidated.4–6 Detection of circulating nucleic acids including DNA and RNA in the plasma and serum of cancer patients, which has genetic characteristics identical to those of the primary tumor, is an emerging field for noninvasive molecular diagnosis of cancers and is attracting considerable interest.7–9 According to the present data, it might be possible to detect over 80% of patients with cancers using a combination of appropriate circulating RNA and DNA markers. Most of the detection of cancer biomarkers focused on the detection of proteins or nucleic acids independently. Research has demonstrated that the combination of protein and nucleic acid markers could be useful for the diagnosis of early-stage diseases and improving the sensitivity and specificity of diagnosis.10,11
Early technologies and platforms for simultaneous detection of nucleic acids and proteins (SDNP) were based on complex and expensive instruments (multiparameter cell analysis system)12 or the combination of nucleic acid detection (DNA array, PCR) and protein detection (protein array, ELISA).11,13–15 Recent research is moving forward the development of an integrated platform and tools for rapid, simple and inexpensive SDNP.16–19 For example, Ladd et al. reported a hybrid surface platform for SDNP using a surface plasmon resonance (SPR) imaging sensor.16 By using DNA-directed protein immobilization on only some of the spots of a DNA array, a mixed DNA/protein array was constructed. Harper et al. described an electrochemical approach for SDNP involving the selective immobilization of DNA and antibody probes on electrode arrays.17 Gabl et al. developed a novel integrated biosensor technology based on thin-film bulk acoustic wave resonators on silicon for SDNP without using a label.18 Shin et al. reported a field effect transistor (FET)-type biosensor based on 0.5 mm standard complementary metal oxide semiconductor (CMOS) technology, and its feasibility for SDNP was investigated.19 However, most of these integrated bioassays are performed in the batch platform and have not been applied for routine use in research laboratories or for clinical diagnosis applications because of the expensive instruments required, reproducibility shortcomings or complex operations, such as multiple incubation and washing steps. There is, therefore, a need for the development of an inexpensive, quick and simple tool with high sensitivity and specificity for SDNP.
Recently, research has concentrated on the development of point-of-care (POC) biosensors for clinical diagnosis applications.20 Emerging lateral flow strip biosensors, also called immunochromotographic test strips, dipstick test strips or dry reagent strip biosensors (DRSB), have been used widely for POC detection of proteins.21–26 The DRSB offers a promising approach to realize POC detection of proteins considering their many advantage such as their user-friendly format, the very short time (generally less than 10 min) to obtain test results, less interference due to chromatographic separation, long-term stability over a wide range of climates, and relatively low cost.21,26 The concept has been expanded by us27–31 and other groups32–36 to develop nucleic acid DRSBs, which avoids multiple incubation, separation and washing steps in the conventional nucleic acid biosensors. In this work, we report a simple and fast method based on the lateral flow strip technology and gold nanoparticles (GNPs) labels for SDNP. The proof of principle was demonstrated by using 60-mer DNA and rabbit IgG (R-IgG) model targets. Qualitative judgment can be performed by observing the color changes of the test lines and quantitative detection can be realized by recording the intensities of the test lines with a portable “strip reader” instrument. The total assay time for a sample containing target DNA and R-IgG is 15 min. The promising properties of the biosensor are reported in the following sections.
Experimental
Reagents and apparatus
Polyester backing materials, nitrocellulose membrane (AE 98), glass fibers, and absorbent materials were purchased from Millipore Corp. (Bedford, MA). Polyclonal goat anti-rabbit IgG and R-IgG were purchased from Pierce Biotechnology (Rockford, IL). HAuCl4, sodium citrate, bovin serum albumin (BSA), sucrose, Triton X-100 and Tween-20, streptavidin from streptomyces avidin, dithiothreitol (DTT), sodium chloride-sodium citrate buffer (SSC, pH 7.0, 20 times concentrated), and phosphate buffer saline (PBS, pH 7.4, 0.01 M) were purchased from Sigma-Aldrich (St. Louis, MO). The SSC buffers with different concentrations were prepared by diluting the concentrated SSC. All chemicals used in this study were analytical reagent grade. All stock solutions were prepared using deionized water purified with the Nanopure System (Barnstead, Kirkland, WA). Glass fibers (GFCP000800), cellulose fiber sample pads (CFSP001700), laminated cards (HF000MC100) and nitrocellulose membranes (HFB18004 and HFB 24004) were purchased from Millipore (Billerica, MA). DNA oligonucleotides were obtained from Integrated DNA Technologies, Inc. (Coralville, IA) and had the following sequences:
Target DNA:
5′-TTCCCTAGCCCACCCAGTGTGCAAGGGCAGTGAAGA CTTGATTGTACAAAATACGTTTTG-3′
DNA probe 1: 5′-ThioMC6-D/CAA AAC GTA TTT TGT ACA A-3′
DNA probe 2: 5′-CAC TGG GTG GGC TAG GGA A/Biotin/-3′
DNA probe 3: 5′-Biotin/TTG TAC AAA ATA CGT TTT G–3′
Noncomplementary DNA: 5′-ATG GCA TCG CTT AGC TGC CAG TAC ACT GAT TGA AGA CAT CAT AGT GCA GAC AAG CAT ATC-3′
The dispensers Airjet AJQ 3000, Biojet BJQ 3000, and Clamshell Laminator as well as the Guillotine cutting module CM 4000 were from BioDot Inc. (Irvine, CA). Strip reader DT1030 was purchased from Shanghai Goldbio Technology Co., Ltd. (Shanghai, China).
Preparation of GNP-DNA and GNP-Ab conjugates
GNPs with average diameter 15 ± 3.5 nm were prepared according to the reported method with slight modifications.27,36 A thiolated oligonucleotide (DNA probe 1) was used for conjugation with GNPs. The GNP–Ab conjugate was prepared according to the reported methods with slight modifications (see details in Supporting Information).27
Preparation of lateral flow device (LFD)
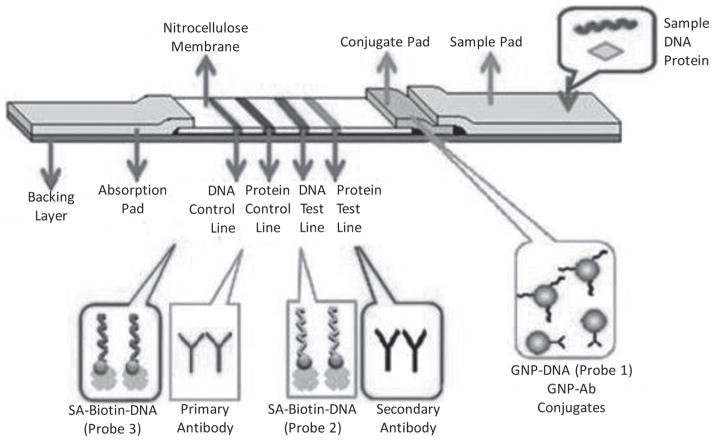
A schematic diagram of the LFD is shown in Fig. 1. The LFD consisted of four components: sample application pad, conjugate pad loaded with the mixture of GNP–Ab and GNP–DNA conjugates, nitrocellulose membrane and absorbent pad. All of the components were mounted on a common backing layer (typically an inert plastic, e.g., polyester) using the Clamshell Laminator. The sample application pad (17 mm × 30 cm) was made from cellulose fiber (CFSP001700, Millipore) and saturated with a buffer (pH 8.0) containing 0.25% Triton X-100, 0.05 M Tris–HCl and 0.15 M NaCl. Then the pad was dried and stored in a desiccator at room temperature. The conjugate pad (8 mm × 30 cm) was prepared by dispensing a desired volume of GNP–DNA and GNP–Ab mixture solution with a ratio of 3:1 onto the glass fiber pad with the dispenser Airjet AJQ 3000, and then drying it under room temperature. The pad was stored in a desiccator at 4°C. Primary antibody, capture DNA probe (DNA probe 2), the secondary antibody (against the Ab on the GNP surface), and control DNA probe (DNA probe 3) solutions were dispensed on the nitrocellulose membrane (25 mm × 30 cm) at different zones to form test lines and control lines, respectively. To facilitate the immobilization of DNA probes on the nitrocellulose membrane, streptavidin was used to react with the biotinylated DNA probes (DNA probe 2 and 3) to form the streptavidin–biotin DNA conjugates. A volume of 60 μL biotinylated DNA probe 2 or probe 3 (1 mM) was added to 250 μL 2.5 mg mL−1 streptavidin solution and incubated for 1.5 h at room temperature. The mixture solution was then transferred to a dialysis filter (cut off value: 30000) to remove excess DNA probes. The streptavidin-biotinylated DNA complexes were dispensed on the nitrocellulose membrane with the dispenser Biojet BJQ 3000. The distance between the test line and control line was around 0.2 cm. The antibody and DNA probes loaded membrane was then dried at room temperature for 1 h and stored at 4°C in a dry state. Finally, the sample pad, conjugate pad, nitrocellulose membrane, and absorption pad were assembled on a plastic adhesive backing (60 mm × 30 cm) using the clamshell laminator as shown in Fig. 1. Each part overlapped 2 mm to ensure the solution would pass through the strip during the assay. Strips with a 4.7-mm width were cut by using the Guillotin cutting module CM 4000.
Fig. 1.
Schematic diagram of the lateral flow device based on the antibody- and DNA-functionalized gold nanoparticle probes for simultaneous detection of nucleic acid and protein. SA: streptavidin.
Sample assay procedure
One hundred thirty microliters of sample solution containing a desired concentration of target DNA and protein in 10× SSC buffer was applied to the sample application zone. After waiting for 10 min, another 40 μL 10× SSC buffer was applied to wash the LFD. This additional washing step would reduce the background of the assay. The bands were visualized within 5 min. For quantitative assays, the LFD was inserted into the strip reader DT1030; the intensities of the red bands (two test lines and two control lines) could be obtained simultaneously by using the portable “Strip Reader” and corresponding software. In the case of the detection of DNA and protein sample in plasma, 2 μL of plasma sample spiked with desired concentrations of DNA and protein was first applied to the sample pad, then 130 μL of 10× SSC buffer was added immediately. Following a washing step (applying 40 μL 10× SSC buffer), the intensities of test lines were read with the strip reader.
Results and Discussion
Principle of the LFD for simultaneous detection of nucleic acid and protein (SDNP)
The LFD (Fig. 1) consisted of four components: sample application pad, conjugate pad, nitrocellulose membrane and absorbent pad. The principle of SDNP with LFD is based on simultaneous sandwich-type DNA hybridization and antibody–antigen immunoreactions. Our previous experiments indicated that the strip biosensor adopting polyclonal antibody as a capture probe and at the same time as a reporter probe showed great performance.37 Polyclonal goat anti rabbit IgG and R-IgG were thus selected as the antibody–antigen pair and a single stranded DNA with 60 bases was selected as a target DNA to demonstrate the SDNP proof of concept on the LFD. In this study, GNPs were used as tags to label antibody (Ab) and DNA reporter probe (DNA probe 1), respectively. The mixture of GNP–Ab and GNP–DNA conjugates were dispensed on the conjugate pad. Capture DNA (DNA probe 2), control DNA probe (DNA probe 3, which is complementary to the reporter DNA probe 1 on the GNP surface), primary antibody (anti-IgG) and the secondary antibody (against anti-IgG) were dispensed on different locations of the nitrocellulose membrane to form two test zones and two control zones, respectively. The sample solution containing the target DNA and protein (R-IgG) is applied on the sample application pad. The solution migrates by capillary action and passes the conjugate pad, and then rehydrates the GNP–DNA and GNP–Ab conjugates. The target DNA hybridizes with DNA reporter probe 1 on the GNP–DNA conjugates to form GNP–DNA–DNA hybrids, and R-IgG (target protein) binds with the GNP–Ab to form GNP–Ab–R-IgG complexes. The complexes and excess of conjugates continue to migrate along the strip. The formed GNP–DNA–DNA and GNP–Ab–R-IgG complexes are captured on the test zones by the second hybridization events (between the target DNA and DNA probe 2 on the test zone) and the second immunoreactions, respectively. The accumulations of GNPs in the test zones are visualized as characteristic red bands (test lines). The excess of conjugates continue to migrate and pass the control zones. Then the excess of GNP–DNA and GNP–Ab are captured by the control DNA probe (DNA probe 3) and secondary antibody in the control zones, respectively, thus forming another two red bands (control lines). In the absence of target DNA and R-IgG, no red bands are observed in the test zones. In this case, two red control bands show that the LFD is working properly. Qualitative analysis is simply performed by observing the color change of the test zones, and quantitative analysis is realized by reading the intensities of the red bands on the test zones with a portable strip reader.
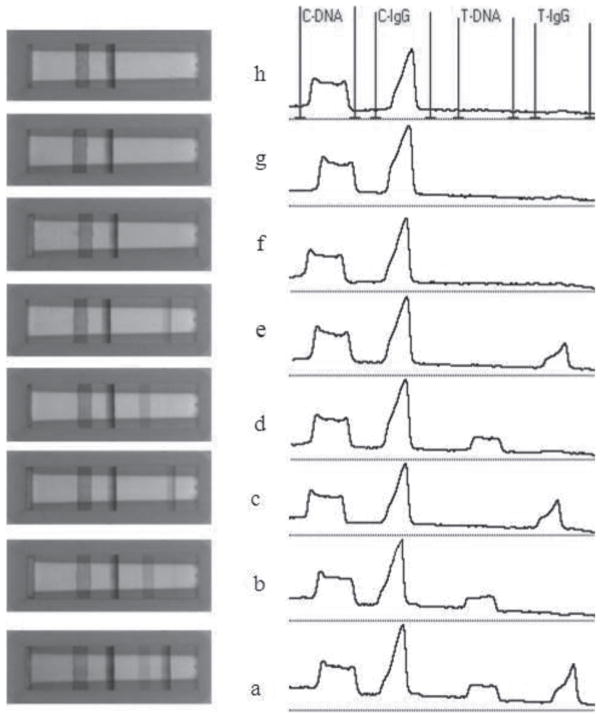
Figure 2 displays the typical photo images and corresponding optical responses of the LFD after applying different sample solutions. As expected, four red bands were observed with the sample solution containing 100 nM target DNA and 100 ng mL−1 R-IgG (Fig. 2a, left). The left two bands are control lines and the right two bands are test lines. Four distinct and resolved peaks were obtained with the strip reader by scanning the bands. The peak shape differs in the formed DNA bands (flat, wide lines) and protein bands (sharp, narrow lines). Such differences would be caused by the different band width. The reason the DNA bands are wider than the protein bands is that the immunoreaction rate is faster than the DNA hybridization reaction rate. The individual target (only DNA, Fig. 2b or only R-IgG, Fig. 2c) yields three bands (one test line and two control lines). No signal was observed in the test zones for neither the control sample (without either DNA or R-IgG, Fig. 2f), as well as in presence of a large excess (10-fold) of noncomplementary DNA (Fig. 2g) or nonspecific protein (BSA, Fig. 2h), reflecting the negligible nonspecific adsorption of the conjugates. Similar responses for individual target DNA (or R-IgG) were observed in the absence and presence of excess of nonspecific protein (or DNA), indicating minimal cross interferences (Figs. 2d and 2e). Other proteins including goat anti mouse IgG and human IgG were also checked. Experiments indicated that the interferences in the presence of an excess of such proteins were too small, so it can be ignored.
Fig. 2.
Typical photo images (left) and corresponding optical responses (right) of the red bands (control lines and test lines) on the LFD after applying different sample solutions. The responses were obtained by reading the intensities of the red bands on the test zones and control zones with a portable strip reader. The concentrations of DNA and IgG in the sample solutions are: (a) 100 nM complementary DNA + 100 ng mL−1 R-IgG, (b)100 nM complementary DNA + 0 ng mL−1 R-IgG, (c) 0 nM complementary DNA + 100 ng mL−1 R-IgG, (d) 100 nM complementary DNA + 0 ng mL−1 R-IgG + 1000 ng mL−1 BSA, (e) 0 nM complementary DNA + 100 ng mL−1 R-IgG + 1000 nM noncomplementary DNA, (f) 1000 ng mL−1 BSA + 0 nM complementary DNA + 0 ng mL−1 R-IgG, (g) 0 nM complementary DNA + 0 ng mL−1 R-IgG + 1000 nM noncomplementay DNA, (h) 0 nM complementary DNA + 0 ng mL−1 R-IgG. The sample solution was prepared using 10× SSC buffer containing 1% BSA. Volume of the sample solution: 130 μL; assay time: 15 min.
Optimization of experimental conditions
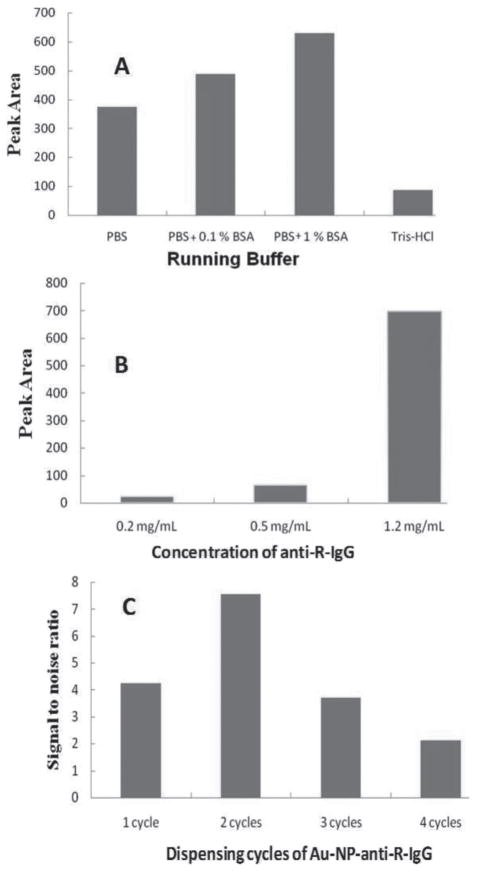
To obtain the best sensitivity and reproducibility of the LFD for SDNP, analytical parameters were optimized. According to our previous report,27 the best conditions for detecting individual DNA target on nucleic acid lateral flow strip biosensor were: six dispensing cycles of the GNP–DNA conjugates on the conjugate pad, 0.12 mM capturing DNA probe dispensed on the test zone and 4× SSC containing 1% BSA buffer. Considering the different kinetics between nucleic acid hybridization and antibody–antigen immunoreactions on the nitrocellulose membrane, the experimental parameters of the LFD for SDNP need to be optimized to obtain the best sensitivity. A tradeoff between the parameters of nucleic acid detection and protein detection with the LFD was made to obtain a similar sensitivity for SDNP. For this purpose, first we prepared the LFD for detecting R-IgG. It was found that the best sensitivity of the LFD for the detection of R-IgG was obtained with two dispensing cycles of GNP–Ab conjugates on the conjugate pad, 1.2 mg mL−1 capturing antibody dispensed on the test zone and PBS containing 1% BSA (Fig. 3). Therefore, 0.12 mM capturing DNA probe and 1.2 mg mL−1 capturing antibody were used to prepare the test zones of the LFD for further optimizations.
Fig. 3.
Optimization of experimental parameters. (A) Effect of the running buffer constituents on the responses of LFD for the detection of R-IgG. Concentration of R-IgG in the sample solution: 100 ng mL−1. The sample solutions were prepared with PBS (0.01 M., pH 7.0) containing 1% BSA. (B) Effect of the concentration of anti-R-IgG dispensed on the test zone on the response of the LFD for the detection of individual R-IgG. (C) Effect of the dispensing cycles of GNP–Ab on the conjugate pad on the response of the LFD for the detection of individual R-IgG. The anti-R-IgG solution with a concentration of 1.2 mg mL−1 was used to prepare the test line. The assay time: 7 min. The volume of the sample solution: 130 μL.
In the current study, the mixture of GNP–DNA and GNP–Ab was used to prepare the conjugate pad. In the individual testing, the optimal dispensing cycles of GNP–DNA and GNP–Ab were six and two, respectively. To obtain similar sensitivity for SDNP with the LFD, the conjugate mixture was prepared by mixing the GNP–DNA and GNP–Ab solution at a ratio of 3:1. It was found that the best signal to noise (S/N) ratios for both DNA and protein tests was obtained by dispensing the conjugate mixture twice on the conjugate pad (results not shown).
Another factor affecting the sensitivity and reproducibility of the LFD for SDNP is the components of the running buffer. Appropriate buffers would minimize the nonspecific adsorption, and increase sensitivity and reproducibility of the LFD. In the current study, we tested the responses of the LFD with SSC and PBS as running buffers for SDNP, and found the SSC buffer produced better response for the simultaneous detection of DNA and protein (results not shown). Thus we optimized the effect of the concentration of SSC on the response of the LFD (Fig. S1, Supporting Information). It can be seen that the S/N ratio of the protein test (blue bar) increases with a decrease in SSC concentration (high dilution times), and the S/N ratio of DNA test (red bar) increases with a decrease in SSC concentration (up until dilution of 10 times), further decrease of the SSC concentration (20×) leads to a decrease of the S/N ratio. The low S/N ratios at high concentrations of SSC for both protein test and DNA test may be caused by the high concentration of salts in SSC buffer. It is well-known that the high concentration of salts is not favorable for antibody–antigen immunoreactions. Although a high concentration of salts benefits DNA–DNA hybridization, this condition is accompanied of a high background, leading to a low S/N ratio. Therefore, 10× SSC buffer was used throughout our experiments.
Performance of the LFD for SDNP
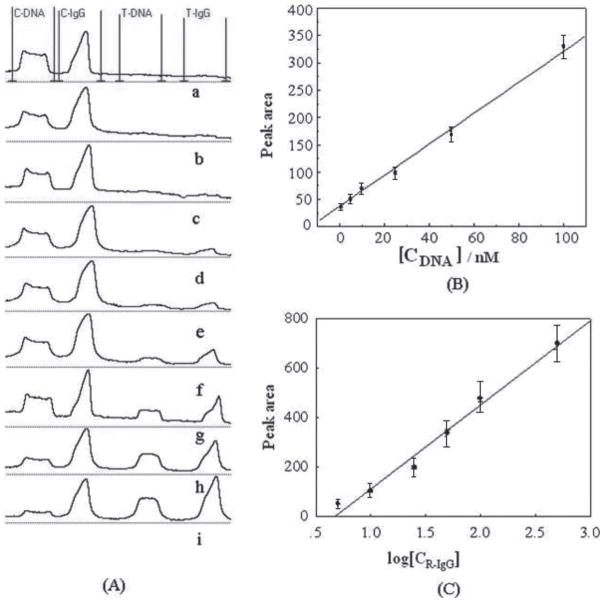
Under optimal experimental conditions, we examined the performance of the LFD with the sample solutions containing different concentrations of target DNA and R-IgG. The size of the peaks and intensities of the test lines reflect the concentration of the corresponding DNA and R-IgG target allowing for convenient qualitative and quantitative assays. Figure 4A displays the recorded responses of the LFD with increasing concentration levels of DNA and R-IgG in the sample solutions. Well-defined peaks with increasing peak areas were observed with an increase in the concentration levels of both DNA and R-IgG. The resulting calibration plot of the peak areas versus DNA concentration (Fig. 4B) is linear over the 1 to 100 nM range and the calibration plot of the peak areas versus logarithm of R-IgG concentration (Fig. 4C) is linear over the 5 to 500 ng mL−1 range. The detection limits of 0.5 nM DNA and 2 ng mL−1 R-IgG (based on S/N = 3) were estimated in connection with a 15 min assay time. Qualitative assay (observed with naked eye) can be used to measure as low as 5 nM DNA and 10 ng mL−1 R-IgG simultaneously. The sensitive and specific response was coupled with high reproducibility. A series of measurements of the mixture of 5 nM DNA and 20 ng mL−1 R-IgG with six LFD yields reproducible signals with RSDs of 4.2 and 3.9%, respectively (data not shown).
Fig. 4.
Analytical performances of the LFD. (A) The typical responses of the LFD with increasing DNA and protein concentrations. From curve a to i, the concentrations of target DNA (nM) and IgG (ng mL−1) are 0, 1, 5, 10, 25, 50, 100, 500, 1000. (B) The resulting calibration curve of DNA test. (C) The resulting calibration curve of R-IgG test. Error bars represent standard deviation, n = 6. Other conditions are the same as in Fig. 2.
Conclusions
In summary, we have developed a simple and fast method based on the DNA probe- and antibody-functionalized gold nanoparticles and lateral flow test strip for simultaneous detection of nucleic acid and protein. A model system comprising IgG and a synthesized single strand DNA target with 60 bases was used to demonstrate the proof-of-concept. Under optimal conditions, the device was capable of detecting a minimum 0.5 nM DNA and 2 ng mL−1 R-IgG simultaneously in 15 min. The device coupled with a portable “strip reader” shows great promise for in-field and point-of-care quantitative/qualitative testing for disease-related circulating nucleic acid and protein biomarkers simultaneously in biological fluids. Considering the fact that real DNA targets in biological fluids have longer sequences in double strands without any pretreatments, the LFD would be more useful for the detection of microRNA and proteins simultaneously. In addition, further improvements in the sensitivity of the device will be required for the detection of real samples. This could be achieved by using enzyme tracer coated gold nanoparticle labels, dye-doped polystyrene nanosphere or silica nanoparticle labels. The concept can be extended for the development of dry-reagent strip biosensor array by immobilizing multiple DNA probes and antibodies on the strip for simultaneous detection of multiple DNA and protein biomarkers. Potential applications of this device for the simultaneous detection of cancer protein biomarkers and microRNA (miRNA) are currently under investigation in our lab. It may offer an innovative tool for early cancer diagnosis, screening and staging of cancer, as well as the evaluation of response to therapy.
Supplementary Material
Acknowledgments
This research was supported by the National Cancer Institute (Grant No. R21CA137703) and the National Institute of General Medicine (NIGMS; Grant No. 5P30 GM103332). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. Y. H. acknowledges financial support from National Science Foundation of China (No. 81201223) and Guangdong Province Medical Research Foundation of China (No. A2012533).
Footnotes
Preparation of GNP–DNA and GNP–Ab conjugates, Fig. S1. This material is available free of charge on the Web at http://www.jsac.or.jp/analsci/.
References
- 1.Ouyang D, Chen J, Getzenberg R, Schoen R. Am J Gastroenterol. 2005;100:1393. doi: 10.1111/j.1572-0241.2005.41427.x. [DOI] [PubMed] [Google Scholar]
- 2.Jian K. Curr Opin Mol Ther. 2007;9:563. [PubMed] [Google Scholar]
- 3.Duffy M. Clin Chem. 2006;52:345. doi: 10.1373/clinchem.2005.059832. [DOI] [PubMed] [Google Scholar]
- 4.Duffy M. Clin Chem. 2001;47:624. [PubMed] [Google Scholar]
- 5.Cheung K, Graves C, Robertson J. Cancer Treat Rev. 2000;26:91. doi: 10.1053/ctrv.1999.0151. [DOI] [PubMed] [Google Scholar]
- 6.Feng Q, Yu M, Kiviat N. Crit Rev Clin Lab Sci. 2006;43:497. doi: 10.1080/10408360600922632. [DOI] [PubMed] [Google Scholar]
- 7.Swarup V, Rajeswari M. FEBS Lett. 2007;581:795. doi: 10.1016/j.febslet.2007.01.051. [DOI] [PubMed] [Google Scholar]
- 8.Anker P, Mulcahy H, Stroun M. Int J Cancer. 2003;103:149. doi: 10.1002/ijc.10791. [DOI] [PubMed] [Google Scholar]
- 9.Bunn P., Jr J Clin Oncol. 2003;21:3891. doi: 10.1200/JCO.2003.07.976. [DOI] [PubMed] [Google Scholar]
- 10.Gygi S, Rochon Y, Franza B, Aebersold R. Mol Cell Biol. 1999;19:1720. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flamini E, Mercatali L, Mercatali L, Nanni O, Calistri D, Nunziatini R, Zoli W, Rosetti P, Gardini N, Lattuneddu A, Verdecchia G, Amadori D. Clin Cancer Res. 2006;12:6985. doi: 10.1158/1078-0432.CCR-06-1931. [DOI] [PubMed] [Google Scholar]
- 12.Crissman H, Steinkamp J. J Cell Biol. 1973;59:766. doi: 10.1083/jcb.59.3.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sansonno D, Cornacchiulo V, Racanelli V, Dammacco F. Cancer. 1997;80:22. [PubMed] [Google Scholar]
- 14.Graham M, II, Bunn P, Jr, Jewett P, Gonzalez-Aller C, Horwitz K. Cancer Res. 1989;49:3934. [PubMed] [Google Scholar]
- 15.Ideker T, Thorsson V, Ranish JV, Christmas R, Buhler J, Eng J, Bumgarner R, Goodlett D, Aebersold R, Hood L. Science. 2001;292:929. doi: 10.1126/science.292.5518.929. [DOI] [PubMed] [Google Scholar]
- 16.Ladd J, Taylor A, Piliarik M, Homola J, Jiang S. Anal Chem. 2008;80:4231. doi: 10.1021/ac800263j. [DOI] [PubMed] [Google Scholar]
- 17.Harper J, Polsky R, Wheeler D, Dirk S, Brozik S. Langmuir. 2007;2:8285. doi: 10.1021/la701775g. [DOI] [PubMed] [Google Scholar]
- 18.Gabl R, Feucht H, Zeininger H, Eckstein G, Schreiter M, Primig R, Pitzer D, Wersing W. Biosens Bioelectron. 2004;19:615. doi: 10.1016/s0956-5663(03)00259-8. [DOI] [PubMed] [Google Scholar]
- 19.Shin J, Kim D, Park H, Lim G. Electroanalysis. 2004;16:1912. [Google Scholar]
- 20.Soper S, Brown K, Ellington A, Frazier B, Garcia-Manero G, Gau V, Gutman S, Hayes D, Korte B, Landers J, Larson D, Ligler F, Majumdar A, Mascini M, Nolte D, Rosenzweig Z, Wang J, Wilson D. Biosens Bioelectron. 2006;21:1932. doi: 10.1016/j.bios.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 21.Weller M. Fresenius’ J Anal Chem. 2000;366:635. doi: 10.1007/s002160051558. [DOI] [PubMed] [Google Scholar]
- 22.Cuzzubbo, Endy T, Nisalak A, Kalayanarooj S, Vaughn D, Ogata S, Clements D, Devine P. Clin Diagn Lab Immunol. 2001;8:1150. doi: 10.1128/CDLI.8.6.1150-1155.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Plotz CM, Singer JM. Am J Med. 1956;21:888. [PubMed] [Google Scholar]
- 24.Kohn J. Immunology. 1968;15:863. [PMC free article] [PubMed] [Google Scholar]
- 25.Millipore C. A Short Guide: Developing Immunochromatographic Test Strips. Millipore Corp; Bedford, MA: 1996. [Google Scholar]
- 26.Liu G, Lin Y, Wai C, Lin Y. Anal Chem. 2007;79:7644. doi: 10.1021/ac070691i. [DOI] [PubMed] [Google Scholar]
- 27.Mao X, Ma Y, Zhang A, Zeng L, Liu G. Anal Chem. 2009;81:1660. doi: 10.1021/ac8024653. [DOI] [PubMed] [Google Scholar]
- 28.He Y, Zeng K, Zhang S, Gurung A, Baloda M, Zhang X, Liu G. Biosens Bioelectron. 2012;31:310. doi: 10.1016/j.bios.2011.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He Y, Zhang X, Zeng K, Zhang X, Baloda M, Gurung A, Liu G. Biosens Bioelectron. 2011;26:4464. doi: 10.1016/j.bios.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He Y, Zhang S, Zhang X, Baloda M, Gurung A, Xu H, Zhang X, Liu G. Biosens Bioelectron. 2011;26:2018. doi: 10.1016/j.bios.2010.08.079. [DOI] [PubMed] [Google Scholar]
- 31.He Y, Zeng K, Gurung A, Baloda M, Xu H, Zhang X, Liu G. Anal Chem. 2010;82:7169. doi: 10.1021/ac101275s. [DOI] [PubMed] [Google Scholar]
- 32.Baeumner AJ, Pretz J, Fang S. Anal Chem. 2004;76:888. doi: 10.1021/ac034945l. [DOI] [PubMed] [Google Scholar]
- 33.Baeumner AJ, Schlesinger N, Slutzki N, Romano J, Lee E, Montagna R. Anal Chem. 2002;74:1442. doi: 10.1021/ac015675e. [DOI] [PubMed] [Google Scholar]
- 34.Glynou K, Ioannou P, Christopoulos T, Syriopoulou V. Anal Chem. 2003;75:4155. doi: 10.1021/ac034256+. [DOI] [PubMed] [Google Scholar]
- 35.Kalogianni D, Bravou V, Christopoulos T, Ioannou P, Zoumbos N. Nucleic Acids Res. 2007;35:e23. doi: 10.1093/nar/gkl1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carter D, Cary R. Nucleic Acid Res. 2007;35:e74. doi: 10.1093/nar/gkm269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mao X, Baloda M, Gurung A, Lin Y, Liu G. Electrochem Commun. 2008;10:1636. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.