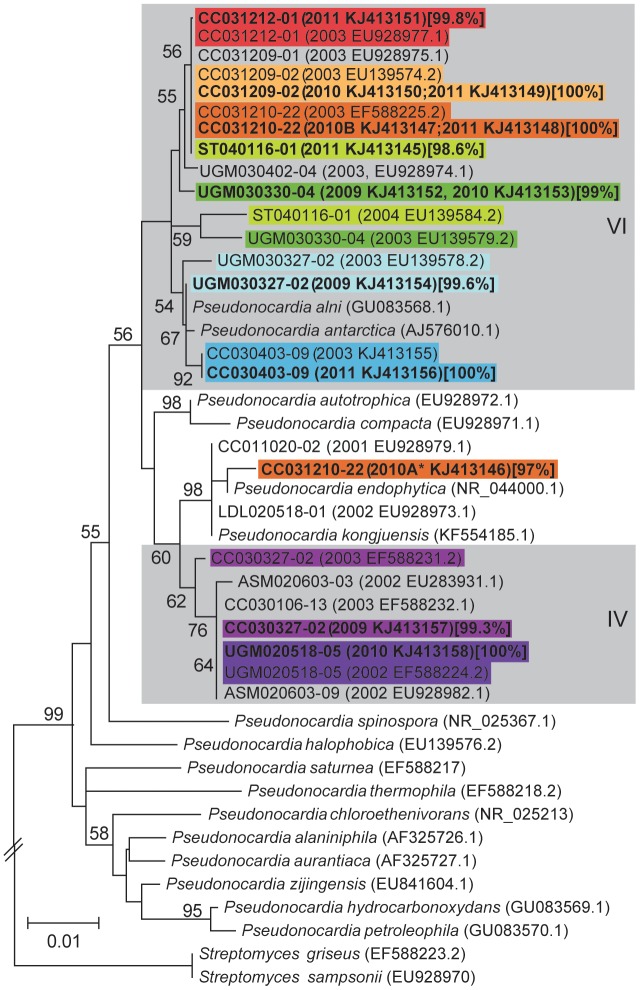
Figure 3. 16S Pseudonocardia phylogenetic tree.
A phylogeny showing bacterial sequences associated with individual leaf-cutting ant colonies over time and their free-living relatives (italics). The phylogenetic tree was generated by the maximum likelihood method based on the Tamura-Nei model and bootstrapped 1000 times. The scale bar represents the genetic distance between samples, reflecting the number of nucleotide changes per site. The tree was created from 16 S rDNA sequences of Pseudonocardia exosymbionts obtained from leaf-cutting ants at the time of colony collection (regular text) and up to 9 years later (bold text). Year(s) of isolation noted in parenthesis after the colony code. Identical sequences obtained at different times are represented by a single sequence with multiple dates. Clades of Pseudonocardia associated with leaf-cutting ants are labeled with roman numerals [8], [19] and depicted in grey boxes. Ant-isolates are labeled by colony number, followed by the year isolated and GenBank identification number in parenthesis. The percentage sequence identity with original isolates is given in brackets after all re-isolates. Isolations from the same colony are boxed in the same color to facilitate comparison. Note that bacteria isolated from all colonies remain in the same phylogenetic clade over time, with the exception of a different morphological type (*) isolated once from a colony with two other independent re-isolates identical to the original sequence isolated.