Figure 5. Hypermethylation of the Fxyd3 promoter reduces transcriptional activity.
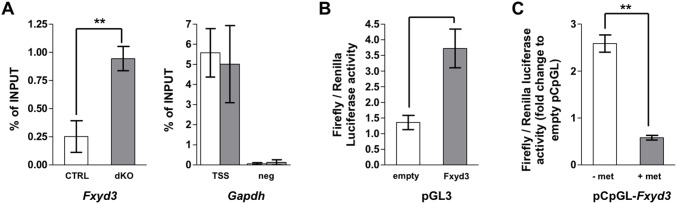
(A) ChIP analysis using H3K4me3 antibody reveals enrichment in this histone mark at the transcriptional start site of the Fxyd3 gene in dKO as compared to control islets. Results are expressed as percent of total input. Data are mean ± sem, n = 3 experiments, *p<0,05. (B) Luciferase activity measured in MIN6 cells transfected with a basic pGL3 or a Fxyd3promoter-PGL3 reporter construct. Data are mean ± sem, n = 7 experiments realized in triplicates, ***p<0,001. (C) The same sequence of the mouse Fxyd3 promoter was sub-cloned into the pCpGL vector (free of CpG). Following in vitro methylation (grey bar) or mock treatment (white bar), basic or Fxyd3promoter-pCpGL plasmids were transfected into MIN6 cells and luciferase activity was measured 48 h later. Methylation significantly reduces luciferase activity. Data are mean ± sem, n = 3 experiments realized in triplicates, **p<0,01. (B, C) Plasmids were co-transfected each time with Renilla reporter vector for normalization.
