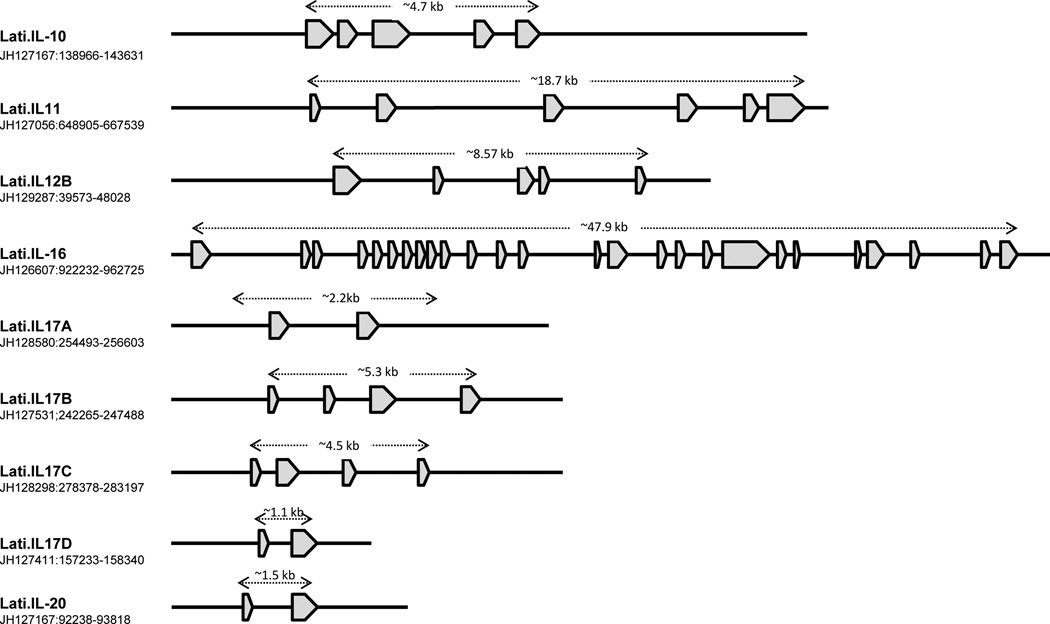
Figure 13. Annotation of the scaffolds encoding coelacanth interleukin genes.
Genes encoding interleukins were identified from the coelacanth annotated assembly at the Ensembl. Coding sequences were retrieved from their corresponding scaffolds and imported into Vector NTI sequence analysis software (Invitrogen). GENESCAN (Burge and Karlin 1997) and BLASTX programs were used, respectively, to predict the exon-intron boundaries and determine amino acid alignments to other vertebrates.