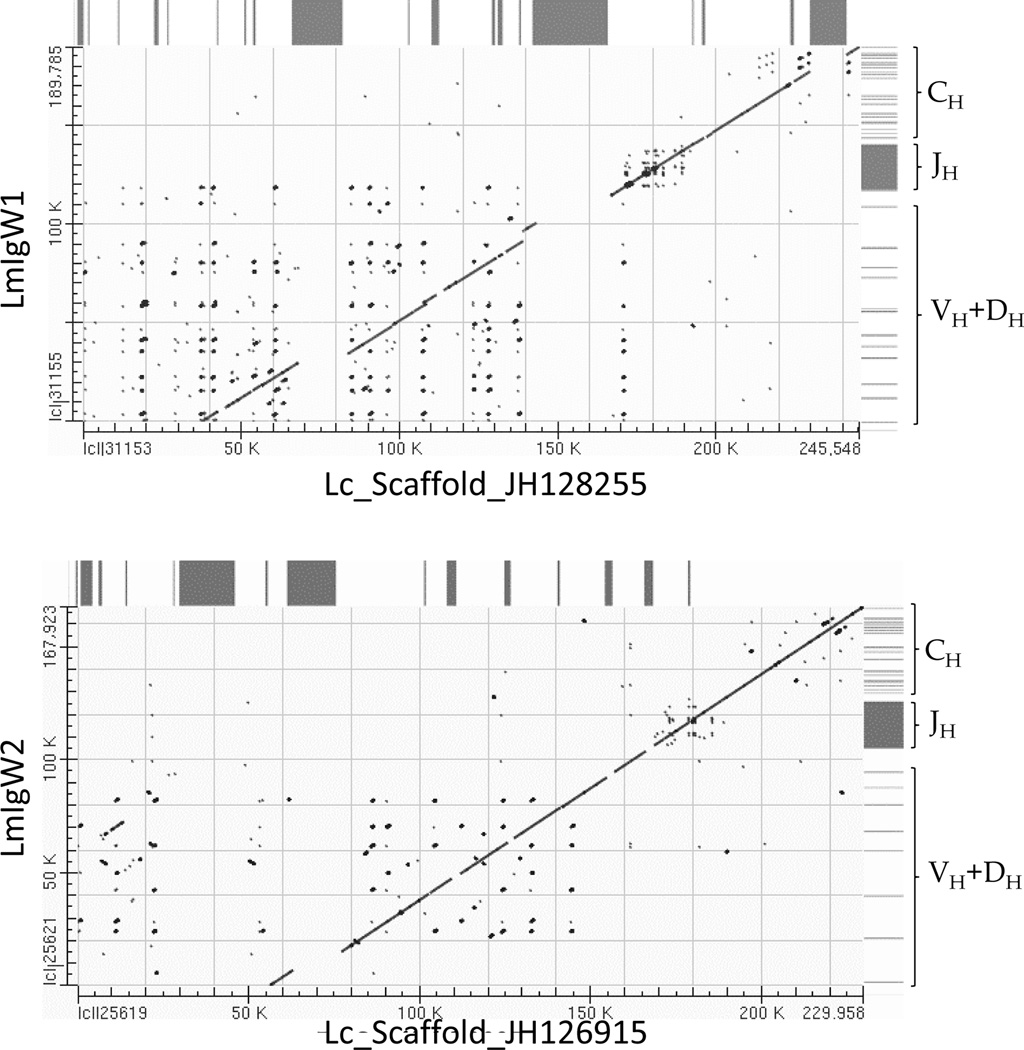
Figure 3. Concordance of L. chalumnae and L. menadoensis genomic sequences encoding two IgW heavy chain loci.
The dot-plots are graphical depictions of MEGABLAST alignments of orthologous IgW heavy chain genomic regions from the two coelacanth species. Horizontal axes represent L. chalumnae scaffolds and vertical axes represent L. menadoensis assemblies based on overlapping BAC clones. Boxes on the right side of the plots denote relative positions of coding sequences in the respective IgW heavy chain loci within the BAC assemblies. Diagonal lines indicate strong concordance between the IgW heavy chain loci of both species. Orthologous regions were highly similar (99% identical across alignable regions). The tracts that are not aligned (gaps in the diagonal line) are largely accounted for by runs of N’s in the L. chalumnae genomic scaffold and are depicted above the plot by black boxes.