Background: Protein fragments of the gap junction Cx43 regulate cellular functions, including resistance to hypoxic stress.
Results: Hypoxia-sensitive IRES activity within the coding region of Cx43 is responsible for generating carboxyl-terminal domains.
Conclusion: Endogenous fragments of Cx43 seem to convey important non-junctional functions.
Significance: Learning how fragments of gap junction proteins are generated is crucial for understanding their functions.
Keywords: Cell Biology, Connexin, Gap Junction, Hypoxia, Protein Processing, Cell Biology, Connexin43, Gap Junctions, Hypoxia, IRES Activity, Protein Processing
Abstract
Connexin43 (Cx43) is the most abundant gap junction protein in higher vertebrate organisms and has been shown to be involved in junctional and non-junctional functions. In addition to the expression of full-length Cx43, endogenously produced carboxyl-terminal segments of Cx43 have been described and have been suggested to be involved in manifold biological functions, such as hypoxic preconditioning and neuronal migration. Molecular aspects, however, behind the separate generation of carboxyl-terminal segments of Cx43 have remained elusive. Here we report on a mechanism that may play a key role in the separate production of these domains. First, stringent evidence derived from siRNA treatment and specific knockouts revealed significant loss of the low molecular weight fragments of Cx43. By applying a dicistronic vector strategy on transfected cell lines, we were able to identify putative IRES activity (nucleotides 442–637) in the coding region of Cx43, which resides upstream from the nucleotide sequence encoding the carboxyl terminus (nucleotides 637–1149). Functional responsiveness of the endogenous expression of Cx43 fragments to hypoxic/ischemic treatment was evaluated in in vitro and in vivo models, which led to a significant increase of the fastest migrating form (20 kDa) under conditions of metabolic deprivation. By nano-MS spectrometry, we achieved stringent evidence of the identity of the 20-kDa segment as part of the carboxyl-terminal domain of full-length Cx43. Our data prove the existence of endogenously expressed carboxyl-terminal domains, which may serve as valuable tools for further translational application in ischemic disorders.
Introduction
Connexins are ubiquitous transmembrane proteins encoded by about 20 different genes in mice and humans (1). All connexin proteins conform to a similar structural organization, consisting of four transmembrane segments linked by two extracellular loops and three cytoplasmic subdomains in the form of the amino terminus, the carboxyl terminus, and a hinge side connecting the second and third transmembrane segment. Among the cytoplasmic portions, the carboxyl-terminal cytosolic tail represents the most variable (in both length and composition) segment of connexins, whereas the amino-terminal domain, capable of self-assembly and hexamer formation, is conserved between members of the connexin family (2). The cytoplasmic C-tail of connexins has been found to be dispensable for channel formation, but it constitutes a site of multiple protein-protein interactions, including phosphorylation through PKA and PKC and anchoring through scaffolding proteins (3, 4).
Cx43, besides conveying channel forming properties, has also been described as being involved in the regulation of additional biological functions, such as control of cell growth, cell survival, and differentiation (5, 6); migration of neural precursor and glioma cells (7–9); and neuroprotection after hypoxic stress (10). An inverse relationship exists between Cx43 expression and cell growth (11, 12), and a carboxyl-terminal domain was found to be as effective as full-length Cx43 in suppressing cell growth in a variety of cell lines (13–15).
Immunoblots of Cx43 usually show three protein bands with a molecular mass in the range of 41–43 kDa. These are thought to represent unphosphorylated and phosphorylated isoforms of Cx43 (16). Interestingly, in most Western blots, higher mobility protein bands of Cx43 (HMBs)2 are also detected with a molecular mass around 20–26 kDa (4). Recently, a single HMB has been suggested to belong to the carboxyl-terminal domain of Cx43 (17), and cardiac ischemic conditions seem to promote the generation of this endogenous CT-Cx43 fragment (4). Keeping in view a possible role of the carboxyl-terminal domain of Cx43 in ischemia, we explored putative molecular mechanisms that may be responsible for the separate generation of carboxyl-terminal domains of Cx43. In this study, we provide evidence that internal IRES activity within the coding region of Cx43 serves a role in internal translation of HMBs. Furthermore, by exploiting chemical metabolic deprivation in a heterologous expression system and in cultured astrocytes, we found that endogenous CT-Cx43 fragments are differentially expressed. This differential expression was further substantiated in an in vivo model, which indicated a substantial increase of CT-Cx43 fragments after hypoxic/ischemic treatment of brain tissue in postpartial rats. Based on these observations, we postulate that a putative IRES activity seems to play a significant role in this mechanism.
EXPERIMENTAL PROCEDURES
Plasmid Constructions
A cDNA clone of rat Cx43-EGFP in a pEGFP-N3 vector was used as template for PCR amplification. DNA fragments from nt 1 to 637 (relative to the translational start of rat Cx43) was PCR-amplified using a sense primer (5′-CGG AAT TCG GTG ACT GGA GTG CCT TGG G-3′), which contains an EcoRI site at the 5′-end, and an antisense primer (5′-GGC TCG AGG AAG ATG ATG AAG ATG GTT TTC TCC G-3′) having an XhoI site at the 5′-end. A PCR product of ∼637 bp was separated on a 1.2% agarose gel and gel-purified using a Qiaex-II gel extraction kit (Qiagen). After digestion with EcoRI and XhoI (Fermentas), this fragment was ligated into the EcoRI/XhoI intercistronic region of the pRF-Di-cis vector (18) to obtain the pRF-1–637 (R1) construct. Similarly, a DNA fragment from nt 1 to 442 was PCR-amplified using the above sense primer and the antisense primer 5′-GGC TCG AGC ATT TTC ACC TTG CCG TGC TC-3′, having an XhoI restriction site at the 5′-end. A PCR product of ∼442 bp was separated on a 1.2% agarose gel, purified, and ligated into the EcoRI/XhoI intercistronic region of the pRF Di-cis vector to obtain the pRF-1–442 (R2) construct. Furthermore, a more downstream DNA fragment from nt 442 to 637 was PCR-amplified using a sense primer (5′-CGG AAT TCA GGG GCG GCT GGC TGA GA-3′) having a EcoRI restriction site at the 5′-end together with the same antisense primer as in the case of the pRF-R1 construct. A PCR product of ∼195 bp was separated on a 1.2% agarose gel, purified, and ligated into the EcoRI/XhoI intercistronic region of the pRF-Di-cis vector to obtain the pRF-442–637 (R3) construct. To replace the firefly luciferase gene with the EGFP gene from the above three Di-cis constructs, the EGFP coding region was obtained from the pEGFP-N3 vector by digesting it with XhoI and NotI. After agarose gel purification, it was ligated at the XhoI and NotI site of the nt 1–637, 1–442, and 442–637 constructs, to get pR-EGFP nt 1–637 (pRG-R1), pR-EGFP nt 1–442 (pRG-R2), and pR-EGFP nt 442–637 (pRG-R3) Di-cis constructs. Promoterless constructs were obtained by removing the CMV promoter from the Di-cis constructs as described previously (30).
The carboxyl-terminal domain of Cx43 (nt 637–1149) was PCR-amplified using the sense primer (5′-CGG AAT TCA TGC TGG TGG TGT CCT TGG) having an EcoRI site at the 5′-end and an antisense primer (5′-TTA AAT CTC CGA GTC ATC AGG-3′) containing a NotI site at the 5′-end. A PCR product of ∼512 bp was separated on a 1.2% agarose gel and gel-purified using the Qiaex-II gel extraction kit (Qiagen). After digestion with EcoRI and NotI (Fermentas), this fragment was ligated into the pEGFP-N3 vector to obtain the CT-Cx43. A deletion construct (deletion of nt 638–714) of Cx43 was prepared by digesting the F712 Cx43 construct (see above) with EcoRV (Fermentas) and EcoRI (Fermentas) restriction enzymes. The double-digested F712 construct was agarose gel-purified from the 712-bp released amino-terminal fragment. The 638-bp amino-terminal Cx43 DNA fragment was PCR-amplified using the sense primer (5′-CGG AAT TCA TGG GTG ACT GGA GTG CCT TGG G-3′), which possessed an EcoRI site at the 5′-end and the antisense primer (5′-TGA AGA TGA TGA AGA TGG TTT TCT CCG-3′). A PCR product of 638 bp was separated on a 1.2% agarose gel and gel-purified using the Qiaex-II gel extraction kit (Qiagen). After digestion with EcoRI (Fermentas) and keeping the 3′- end blunt, this fragment was ligated into the EcoRI/EcoRV-digested F714 construct. The resultant construct of Cx43 lacked the DNA sequence from 638 to 714 bp while keeping the rest in frame.
Frameshift Experiments
A rat Cx43 cDNA clone in pEGFP-N3 vector (without the EGFP gene) was used for mutation studies. A frameshift mutation (F590) was introduced at position nt 590 of the rat Cx43 protein coding region by the addition of a single nucleotide, T, between positions nt 590 and 591 using the full-length Cx43 construct. The mutagenic primer used was 5′-ACC AGG TAG ATC TGC TTC CTC-3′, with the nucleotide underlined. Eventually, the addition of T resulted in the creation of a BglII restriction site (used for screening the mutants). A further frameshift mutation (F712) was created at position nt 712 by the addition of two nucleotides, AT. The mutagenic primer used was 5′-CAA AGG CGT TAA GGA TAT CGC GTG AAG GG-3′, with the nucleotides underlined. The addition of AT resulted in the creation of an EcoRV restriction site (used for screening the mutants).
Cell Culture, Transfection, and Reporter Assays
N2A and NIH3T3 cells were purchased and maintained in cell culture as recommended by the ATCC (LGC Promochem GmbH, Wesel, Germany). For determination of IRES activity, 2 × 104 NIH3T3 cells were plated in 96-well flat bottom plates (BD Biosciences) and transiently transfected using 100 ng of plasmid DNA and the Effectene® transfection protocol (Qiagen, Hilden, Germany). 48 h after transfection, luciferase activity was measured in an Orion II microplate luminometer (Berthold Detection Systems, Pforzheim, Germany), using the Dual-Luciferase reporter assay system (Promega Corp., Madison, WI). IRES activity was expressed as the ratio of firefly luciferase/Renilla luciferase (FLuc/RLuc) with the activity of the control vector (pRF Di-cis) set to 1. Each experiment was performed five times with all constructs in triplicates (changes are indicated throughout). Data are expressed as mean ± S.E. Cultures of primary astrocytes were obtained from P0–P2 wild type mice and astrocyte-directed conditionally deficient Cx43 (Cxfl/fl:Cre) and Cx43/Cx30 conditional double (Cxfl/fl:Cre) mice (19, 20). Astroglial cultures were maintained as described in detail elsewhere (21). All experiments were approved by legal authorities (Landesamt für Natur, Umwelt, und Verbraucherschutz NRW, Germany, record token AZ 50.8735.1) and conformed with national regulations.
RNA Expression Analysis
For expression analysis, 3 × 105 N2A cells were seeded in 24-well plates (BD Biosciences). RT-PCR was performed as a test for mRNA splicing in NIH3T3 cells following DNA transfection of the IRES containing Di-cis constructs, pRF-R1 (R1), pRF-R2 (R2), and pRF-R3 (R3). Total RNA was isolated as described above from N2A cells 48 h after transfection. First strand cDNA synthesis was carried out as described. 18 S rRNA with 18 S sense (5′-catg gtg acc acg ggt gac-3′) and 18 S antisense (5′-ttc ctt gga tgt ggt agc cg-3′) served as internal controls. Construct-specific cDNAs were PCR-amplified using forward primer D1 (5′-GCT TCT GAC ACA ACA GTC TCG AAC TTA AG-3′; upstream of an intron in pRF-Di-cis vector) and reverse primer D2 (5′-CTT CCA GCG GAT AGA ATG GCG CCG GGC C-3′; corresponding to the coding region of firefly luciferase).
Western Blot Analysis
For Western blot analysis of transfected cell lines, 105 NIH3T3 or 2 × 105 N2A cells were grown in 12-well plates (BD Biosciences). Transient transfections were performed using 300 ng of plasmid DNA and the Effectene® transfection protocol (Qiagen). 48 h after transfection, whole cell extracts were prepared according to the manufacturer's protocol (Active Motif nuclear extraction kit, Rixensart, Belgium). 20 μg of each protein fraction was separated on 12% SDS-PAGE and processed as described previously (22). Primary antibodies used were as follows: rabbit polyclonal anti-Cx43, 1:1000 (Zymed Laboratories Inc., South San Francisco, CA); mouse monoclonal anti-GFP, 1:2000 (Roche Applied Science); mouse anti-Renilla luciferase monoclonal antibody, 1:2000 (Millipore, Billerica, MA); and mouse monoclonal anti-β actin antibody, 1:10,000 (Sigma-Aldrich). Peroxidase-labeled anti-mouse IgG (Jackson ImmunoResearch, Europe Ltd.; 1:7,500) and anti-rabbit IgG (Jackson ImmunoResearch; 1:2000) were used as secondary antibodies. In some experiments, the detection antibody was replaced, and the IRDye 680 goat anti-mouse IgG (1:20,000) and IRDye 800 goat anti-rabbit IgG (1:10,000) detection antibodies were used in combination with the Odyssey infrared detection system (LI-COR Biosciences, Lincoln, NE) to quantify protein expression. Quantification of the protein bands was performed using the LI-COR software package. Western blots were prepared from either 12% SDS-PAGE or from 15% SDS-PAGE, as indicated in the respective figure legends. 15% SDS was used when better separation of the low molecular weight proteins was required (i.e. for protein quantification in whole brain samples).
Statistical analysis for Western blot quantification was performed as a one-way analysis of variance (ANOVA) for n = 4 for each of the respective groups. The number of asterisks indicates the level of significance as follows: *, p < 0.05; **, p < 0.01; ***, p < 0.001.
HA-Cx43 Plasmid Construction
Amino-terminal tagged HA-43 construct was generated in mammalian expression vector, pCMV-HA, by PCR-amplifying Cx43 cDNA using pCMV-Cx43 vector as template. For this purpose, forward primer with an EcoRI restriction site (5′-GCC GAA TTC AGG GTG ACT GGA GTG CCT TGG GGA AAG-3′) and reverse primer with a BamHI site (5′-GCA CTC GAG TTA AAT CTC CAG GTC ATC AGG CCG AGG-3′) were used to PCR-amplify Cx43 cDNA. After double digestion with EcoRI/BamHI restriction enzymes (New England Biolabs), the Cx43 cDNA was gel-purified and ligated at EcoRI/BamHI-digested pCMV-HA vector to obtain the HA-Cx43 fusion construct. For Western blotting, the HA-Cx43 construct was transiently transfected in NIH3T3 cells, and 48 h post-transfection, protein extract was prepared and loaded on 15% SDS-PAGE. Western blotting was performed using rabbit anti-HA (1:2000) (Sigma) as a primary antibody. Secondary detection was performed as described elsewhere in this work (see above).
Phosphatase Treatment
For phosphatase treatment, the NIH3T3 cell extract was treated with 1.5 units of potato acid phosphatase (Sigma-Aldrich) in PIPES buffer (20 mm PIPES, 20 mm KCl, 1 mm DTT, 1 mm MgCl, pH 4.8) for 60 min at 37 °C in a total reaction volume of 100 ml. The phosphatase-treated protein extract was used for Western blotting using anti-Cx43 antibody. For control studies, NIH3T3 cell extract was treated as above, omitting phosphatase enzyme from the reaction buffer. After incubation, 100 μg of protein was loaded on 15% SDS-PAGE, and Western blotting was performed using polyclonal anti-Cx43 antibody and further processed as described above.
Metabolic and Ischemic Deprivation
For induction of chronic intermittent hypoxia (CIH) in NIH3T3 cells and mouse primary astrocytes, cells were incubated for 15 min in Hanks' balanced salt solution at 37 °C, followed by treatment with 0.3 mm sodium iodoacetate (Sigma) and 1 mm sodium cyanide (Sigma) for different times (0, 45, and 90 min) at 37 °C (23). Post CIH treatment, cells were sampled, and whole cell lysates were prepared as described previously (30). Western blots were performed as described above. Astrocytes were obtained from postnatal day 2 mouse pups as described (21) and cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. Then cells were plated into 100-mm Petri dishes (Fisher) and allowed to grow for 24 h until the culture medium was changed. All cultured cells were maintained in this medium, which was changed three times a week. Astrocytes were cultured for 2–3 weeks and passaged twice before usage.
For in vivo induction of ischemia/hypoxia, postnatal rats at the age of 48 days were deeply anesthetized by inhalation of 4% halothane and maintained with 1.5% halothane in 50% N2 O, 50% O2. The left common carotid artery was exposed, double-ligated with 6-0 surgical silk, and severed. The duration of anesthesia and surgery did not exceed 10 min. After surgery, the animals were allowed to recover in their home cages for 1 h. To induce systemic hypoxia, the animals were subsequently placed in an incubator (Incubator 7511, Draeger, Germany) and exposed to a hypoxic gas mixture (8% oxygen, 92% nitrogen) for 80 min. The environmental temperature was strictly maintained at 21 °C. All surgical and experimental protocols were approved by the institutional review committee (token AZ 50.8735.1). Approval met the guidelines of the German animal protection law.
Mass Spectrometry
Cultures of neonatal rat primary astrocytes were treated according to ischemic/hypoxic conditions for 90 min as described above. Cell extracts were separated via 12% SDS-PAGE with subsequent visualization of proteins by fast SimplyBlueTM SafeStain (Invitrogen), and stained gel bands were cut out and followed by tryptic in-gel digestion.
The nano-HPLC analysis was performed on an UltiMate 3000 RSLCnano LC system (Dionex, Idstein, Germany). Samples were loaded on a trap column (Thermo; 100 μm × 2 cm, particle size 5 μm, pore size 100 Å, C18) with a flow rate of 30 μl/min with 0.1% TFA. After washing, the trap column was serially connected with an analytical C18 column (Thermo, 75 μm × 50 cm, particle size 2 μm, pore size 100 Å). The peptides were separated with a flow rate of 400 nl/min using the following solvent system: 0.1% formic acid (A); 84% acetonitrile, 0.1% formic acid (B). The separation gradient was from 4 to 40% B for 95 min, and then a washing step was performed at 95% B (for 5 min), and an equilibration step from 95 to 4% B was applied. The HPLC system was connected on-line to the nanoelectrospray ionization source of a Q Exactive mass spectrometer (Thermo Fisher Scientific). In the electrospray ionization-MS/MS analysis, full MS spectra were scanned between 350 and 1400 m/z with a resolution of 70,000 at 200 m/z (AGC target 3e6, 80-ms maximum injection time). The spray voltage was 1600 V (+), and the capillary temperature was 250 °C. Lock mass polydimethylcyclosiloxane (m/z 445.120) was used for internal recalibration. The m/z values initiating MS/MS were set on a dynamic exclusion list for 30 s, and the 10 most intensive ions (charge +2, +3, +4) were selected.
MS/MS fragments were generated by higher energy collision-induced dissociation, in which ion dissociation was performed at normalized collision energy of 27%, fixed first mass 130.0 m/z, and isolation window 2.2 m/z. The fragments were analyzed in an Orbitrap analyzer with 35,000 resolution at 200 m/z (AGC 1e6, maximum injection time 120 ms).
Automatic spectrum interpretation for peptide identification was done using MASCOTTM V software (Matrixscience, London, UK). As references for protein sequences, UniProt (Uniprot/Swissprot-Release 2013_05 of 01.05.2013; 540,052 entries without decoys) was used. The setting for taxonomy was Rattus norvegicus with 5 ppm for precursor mass tolerance and 20 millimass units as fragment mass tolerance. We performed different searches by changing the number of miss cleavages from one up to two, when the first calculated protein fragment was expected not to match with a tryptic cleavage site.
RESULTS
Immunolabeling of Cx43 Detects High Mobility Bands (HMBs) in Addition to Main Cx43 Protein, Which Disappears after Acute Knockdown and Conditional Knockout
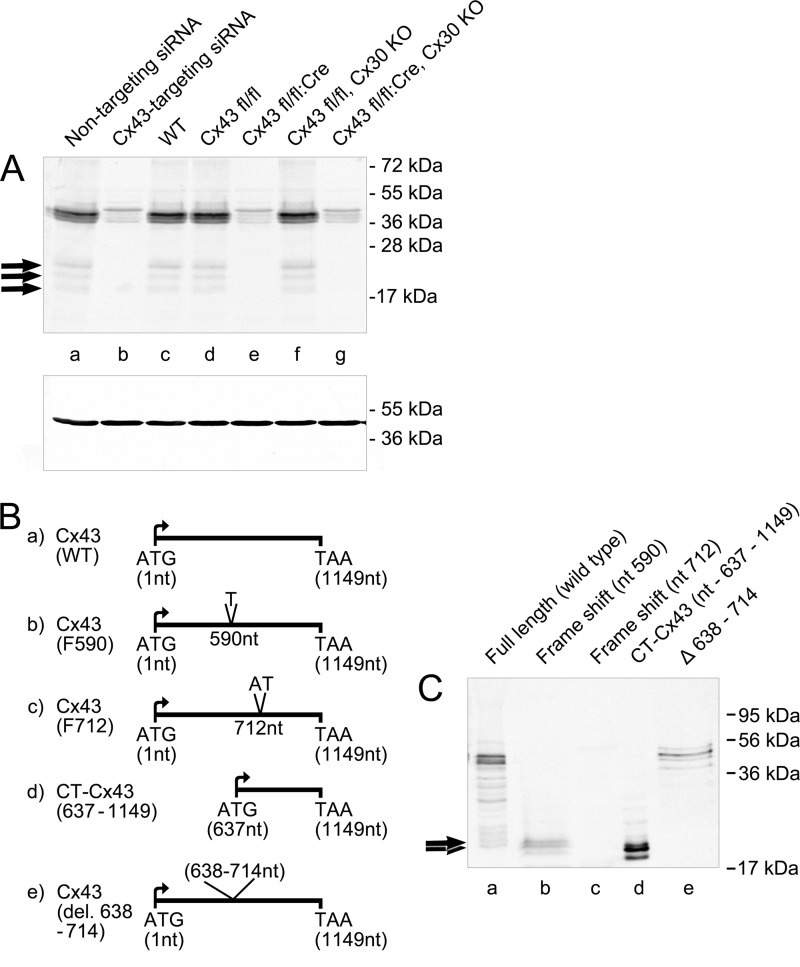
A common notion is that Western blots of Cx43 detect higher HMBs (4, 17) in addition to the main full-length P0–P2 isoforms (16). To validate the existence and specificity of these higher mobility protein bands in our brain samples, total protein extracts were prepared from adult rat brain and rodent astrocytes and run on 12% SDS-PAGE. Western blot detection using a peptide-specific Cx43 antibody (directed toward a carboxyl-terminal peptide of Cx43, amino acids 363–382; Sigma) showed that protein bands around 41–43 kDa, corresponding to the known full-length Cx43 isoforms, were detected in astrocytes (Fig. 1, A, lanes a and c). In addition, HMBs between 20 and 26 kDa were detected (Fig. 1, lanes a, c, d, and f).
FIGURE 1.
A, existence of higher mobility protein band(s) in immunoblots of Cx43. The gel shows full-length bands and HMBs (arrows) of Cx43 from untreated (lane a) and siRNA-treated astrocytes (lane b), astrocytes from wild type mice (WT; lane c), and Cx43 homzygote floxed animals (lane d). Cx43 siRNA (lane b) Cre-activated deletion (Cx43 fl/fl:Cre; lane e) led to a loss of HMBs in astrocytes from transgenic animals, whereas full-length Cx43 isoforms were reduced by about 80%. Cx43 fl/fl animals carrying a transgene with a deleted Cx30 gene, encoding for the second major astrocytic gap junction protein, show persistent expression of Cx43-HMBs and full-length Cx43 (lane f). Deletion of both astroglial connexins (lane g) shows a pattern identical to the Cx43 fl/fl:Cre animals. Protein cleavage is not responsible for the generation of HMBs. B, schematic view of the Cx43 constructs. a, WT full-length Cx43; b, frameshift mutation at nt 590 (F590) exploited for evaluating molecular mechanisms behind the generation of endogenous CT-Cx43 domains; c, frameshift mutation at nt 712 (F712); d, construct expressing the carboxyl-terminal domain (nt 637–1149); e, construct with deletion of nt 638–714. C, Western blot of the above constructs transiently transfected into N2A cells. Lane a, WT Cx43 shows full-length Cx43 and HMBs (arrows). Lane b, frameshift F590 shows a complete loss of full-length Cx43 expression with persistent expression of HMBs (arrows). Frameshift at nt 712 (F712; lane c) displays complete loss of expression of both full-length Cx43 and HMBs. Lane d, expression of the carboxyl-terminal domains after transfection with the CT-Cx43 construct (nt 637–1149). Lane e, deletion of a sequence covering the initial carboxyl-terminal domain (nt 638–714) reveals loss of HMB expression, with persistent expression of full-length Cx43. Variability in the intensity of bands is due to differences in the expression efficiency of the different constructs.
Evidence that the high mobility bands (Fig. 1A, lanes a, c, d, and f) comprise derivatives of the full-length Cx43 was obtained by siRNA silencing of Cx43 in cultured astrocytes, a neural cell type highly enriched in Cx43. Upon exposure of astrocytes to targeted Cx43 siRNA, high mobility bands disappeared in correspondence with an about 80% reduction of the full-length isoforms (Fig. 1A, lane b). In accordance with the responsiveness of Cx43-HMBs to acute knockdown by Cx43 silencing was the complete loss of the HMBs after conditional knockout in astrocytes. Two transgenic mouse strains were exploited to look for response of Cx43-HMBs, when Cx43 was deleted in astrocytes by recombinant cre-loxP technologies (19). Mice lacking Cx43 in astrocytes (Cx43fl/fl:Cre) revealed complete loss of HMBs (Fig. 1A, lane e), unlike the control littermates (Cx43fl/fl; Fig. 1A, lane d). Besides single ablation, conditional double knockouts, including the second major astroglial gap junction protein Cx30 (Cx43fl/fl:Cre,Cx30ko) were also evaluated. As indicated in Fig. 1A, lane g, Western blots of the Cx43 fl/fl:Cre/Cx30ko strain was devoid of HMB immunoreactivity, in contrast to the Cx43fl/fl:Cx30ko mice (Fig. 1A, lane f). These data provide stringent evidence that the HMBs represent part of the Cx43 complement and are in accordance with data on single HMB occurrence, as described recently by Joshi-Mukherjee et al. (17). However, the coordinate disappearance of full-length Cx43 and HMBs does not exclude a putative cleavage of carboxyl-terminal fragments.
Frameshift Experiments Rule Out Possible Proteolytic Cleavage as a Source of CT-Cx43 Fragments
One of several questions raised by the above results is whether a specific molecular mechanism is responsible for the generation of endogenous CT-Cx43 domains. To rule out the possibility of proteolytic cleavage of Cx43, we introduced frameshift mutations. Two mutated cDNA constructs were engineered: one by introducing a single nucleotide, T, between nt 590 and 591 in the coding region of Cx43 cDNA (F590), and a further one (F712) by introducing two nucleotides, AT, between nt 712 and 713 in the coding region of Cx43 (see Fig. 1B for orientation). The frameshift constructs (F590 and F712), and a wild type construct (WT) were transiently transfected into N2A cells (which are known to express no detectable levels of endogenous Cx43). 48 h post-transfection, cell extracts were prepared and separated on 12% SDS-PAGE. Full-length Cx43 and low levels of HMBs were detectable by Western blot of the WT construct. By creating the frameshift at nt 590, translation of full-length Cx43 was completely abolished (Fig. 1C, lane b), whereas the higher mobility bands could still be detected. HMBs showed a similar migration pattern as the carboxyl-terminal domains obtained after transfection with the carboxyl-terminal construct (CT-Cx43, nt 637–1149; Fig. 1C, lane d). Furthermore, the Cx43 construct with a frameshift at position nt 712 resulted in the complete loss of both Cx43 and the carboxyl-terminal domains (Fig. 1C, lane c). The presence of HMBs after the F590 frameshift rules out the possibility of a proteolytic cleavage mechanism responsible for the generation of separate Cx43-CT fragments.
To identify the initiation codon for the generation of CT-Cx43, AUG codons (at positions nt 638, 842, and 959) in the carboxyl-terminal part of Cx43 coding sequence were mutated. However, we were still able to detect HMBs along with full-length Cx43 (data not shown). Because the frameshift experiments indicated that the region between nt 590 and 712 is indispensable for the generation of CT-Cx43, we deleted a DNA sequence (nt 638–714) from the Cx43 construct while keeping the rest in frame. Western blot detection using the CT-specific antibody indicated that deletion of this DNA sequence resulted in the complete loss of HMBs without affecting the expression of full-length Cx43 (Fig. 1C, lane e).
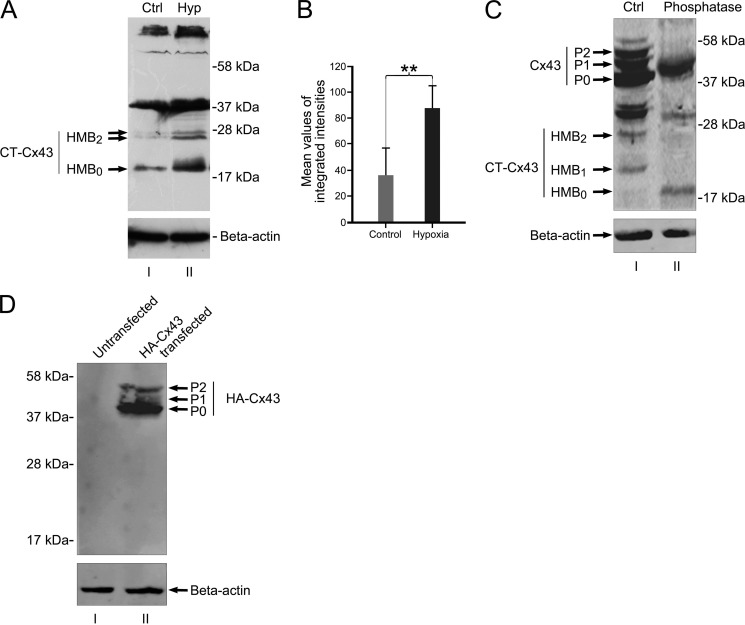
In a final approach to exclude proteolytic cleavage as a possible source of HMBs, we labeled the amino terminus of full Cx43 with an HA tag. If low molecular fragments are generated by proteolytic cleavage of the full-length Cx43, one expects different protein fragments, some of them carrying the HA-tagged amino terminus. This amino-terminal fragment will be detected by HA antibodies, and the CT fragment will remain undetected under Western blot conditions. As shown in Fig. 5B (lane II), HA-tagged fragments were not detectable below the full-length isoforms, which strongly speaks against a proteolytic origin of the HMBs.
FIGURE 5.
In vivo expression of HMBs is responsive to ischemia/hypoxia. Juvenile rats respond with a significant increase of the HMB0 form after ischemia. A, HMBs of brains from untreated animals (lane I) show faint reactions at the height of HMB0 and a doublet band at HMB2, whereas the treated site reveals a robust reaction with dominance of the HMB0 form (lane II). B, mean values of the accumulated protein of all three bands indicate a significant increase (**, p < 0.01) in total protein level after ischemic/hypoxic treatment. n = 4 independent experiments. C, phosphatase treatment confirms that HMB1 and HMB2 are phosphoforms of HMB0. Lane I, protein extract from NIH3T3 cells treated with PIPES buffer, omitting phosphatase enzymes and incubated at 37 °C for 60 min (Ctrl). Lane II, NIH3T3 protein extract treated with potato acid phosphatase and incubated at 37 °C for 60 min. Phosphatase treatment resulted in the loss of P1 and P2 phosphoforms of full-length Cx43 as compared with phosphatase-omitted extract (lane I). Moreover, phosphatase treatment reveals loss of HMB1 and HMB2 and persistence of HMB0. D, Western blot of amino-terminally tagged HA-Cx43 shows no lower fragments of HA-Cx43. Lane I, untransfected NIH3T3 protein extract. Lane II, extract from HA-Cx43-transfected NIH3T3 shows full-length Cx43 isoforms (P0, P1, and P2). Further HA-Cx43 bands in the lower range are not detected by anti-HA antibodies, indicating that protein truncation of full-length Cx43 is unlikely to be responsible for the generation of high mobility bands. Error bars, S.E.
Internal Translation Mediated by a Putative IRES Activity Is a Possible Mechanism for the Separate Expression of Endogenous CT-Cx43
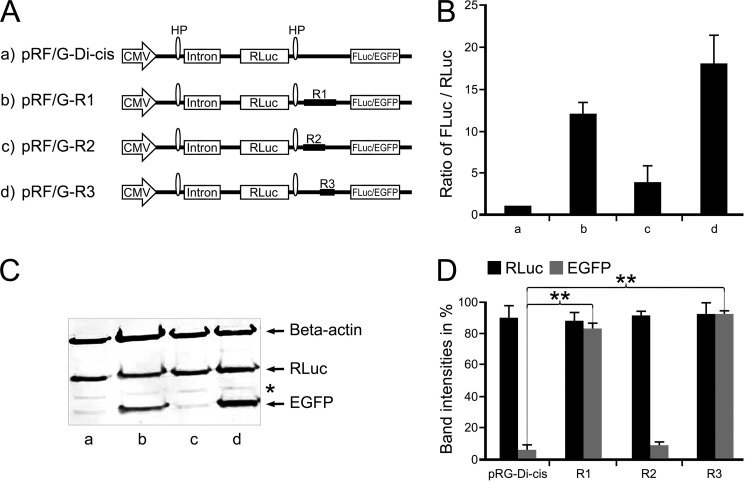
The possibility of an in-frame IRES-mediated mechanism for separate expression of endogenous CT-Cx43 fragments was further evaluated by a dicistronic vector strategy.
For this purpose, DNA fragments from the coding region (R1, nt 1–637; R2, nt 1–442; R3, nt 442–637) of Cx43 were subcloned into the intercistronic region of either a pRF-Di-cis vector (having firefly luciferase as a downstream cistron) or a pRG-Di-cis vector (having EGFP as a downstream cistron) (Fig. 2A). These Di-cis constructs (R1, R2, and R3) along with control Di-cis vector (pRF-Di-cis) were transiently transfected in NIH3T3 cells. Renilla and firefly luciferase activity were measured 48 h after transfection. Luciferase activity readings indicated that the DNA fragment R1 was able to enhance the expression of the downstream firefly luciferase cistron by ∼12-fold (Fig. 2B, column b), DNA fragment R2 enhanced the expression of firefly luciferase by ∼2–3-fold only (Fig. 2B, column c), and the DNA fragment R3 showed a ∼17-fold enhancement of firefly luciferase (Fig. 2B, column d), as compared with control pRF-Di-cis vector (Fig. 2B, column a). IRES activity was expressed as FLuc/RLuc with the activity of the control vector (pRF-Dis-cis) set to 1.
FIGURE 2.
Identification of IRES activity in the coding region of Cx43 using Di-cis vectors. A, schematic view of the Di-cis constructs with Renilla gene as first cistron and either firefly luciferase (FLuc for luciferase data) or EGFP (for Western blot) as second cistron. a, pRF/G-Di-cis (control vector); b, pRF/G-R1 Di-cis construct (R1) (contains coding region from nt 1 to 637; c, pRF/G-R2 Di-cis construct (R2) (contains coding region from nt 1 to 442); d, pRF/G-R3 Di-cis construct (R3) (covering the region from nt 442 to 637). B, IRES activity of the above constructs in NIH3T3 cells. IRES activity is represented as the ratio of firefly to Renilla luciferase luminescence (FLuc/RLuc), with the luminescence of the control vector pRF-Di-cis set at 1. R1 enhances IRES activity by about 12-fold (column b), as compared with vector alone (column a); fragment R2 shows minimal (2–3-fold) luminescence of the FLuc/RLuc ratio (column c). Maximal expression (17-fold) of FLuc was achieved with fragment R3, which contains the nucleotide segment nt 442–637 (column d). C, Western blot of the above Di-cis constructs (with EGFP as second cistron) transiently transfected into NIH3T3 cells. The expression of EGFP corroborates the pattern as found with the dual luciferase assay (shown in B). Fragment R1 (lane b) and fragment R3 (lane d) enhance the expression of the second cistron (EGFP) appreciably, as compared with control vector (lane a) and fragment R2 (lane c). Note that the β-actin protein band (probed in the same gel) represents equal loading of protein samples, and expression of the first cistron (i.e. Renilla luciferase (also probed in the same gel) (RLuc)) remains almost similar in all constructs. D, quantitative assessment of three different Western blots, as shown in Fig. 2C. Each experiment was performed five times with constructs in triplicates. Asterisk, nonspecific protein band. Data are expressed as mean ± S.E. (error bars); **, p < 0.01.
Additional confirmation of the presence of a putative IRES activity was obtained by Western blot analysis. All Di-cis constructs were transiently transfected into NIH3T3 cells, and cell lysates were prepared 48 h after transfection. Immunodetection was performed using an EGFP antibody (Sigma) and anti-RLuc (Millipore) antibodies. As shown in Fig. 2C (lane b), the DNA fragment R1 resulted in a robust enhancement of expression of the downstream cistron EGFP as compared with control vector (Fig. 2C, lane a). DNA fragment R2 showed a low level of expression (Fig. 2C, lane c) almost similar to the control vector, whereas the R3 DNA fragment (nt 442–637) showed the highest expression of EGFP (Fig. 2C, lane d). The expression of Renilla luciferase, RLuc (first cistron), remained similar in all constructs, indicating that the expression of the downstream cistron (EGFP) was mediated by the Cx43 sequence. Quantification of the Western blots corroborated the differential activity of the DNA fragments (Fig. 2D) with a likely localization of the IRES activity in the nt 442–637 region.
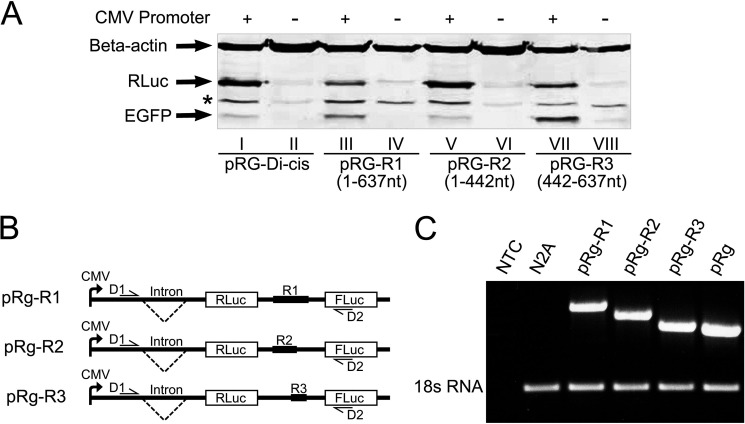
Cryptic Promoter Activity Is Not Present in the Putative IRES Element of Cx43
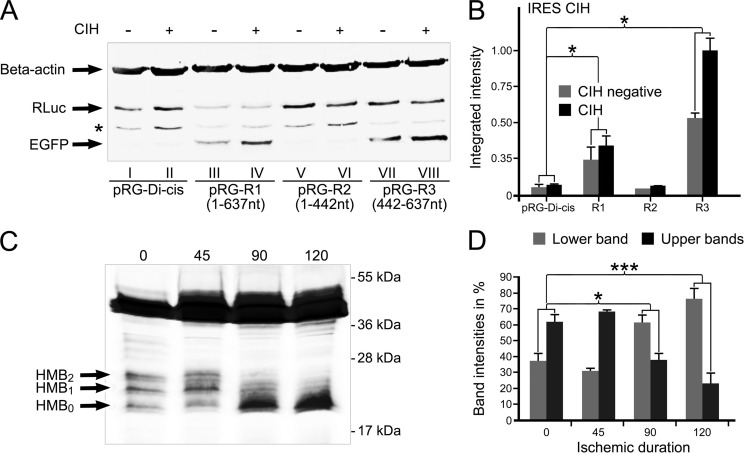
To rule out the possibility that the increased expression of the second cistron in the Di-cis assay is due to cryptic promoter activity, promoterless Di-cis constructs were prepared by removing the CMV promoter (Fig. 3A, lanes II, IV, VI, and VIII). Immunodetection with anti-RLuc and anti-EGFP showed that by removing the CMV promoter from the construct pRG-R1, the expression of the second cistron (EGFP) was almost completely abolished (Fig. 3A, lane IV), whereas the same construct, pRG-R1 (with CMV), showed enhanced expression of EGFP (Fig. 3A, lane III). Furthermore, the most active pRG-R3 construct, when promoterless, also revealed negligible expression of EGFP (Fig. 3A, lane VIII) as compared with the promoter-driven pRG-R3 construct (Fig. 3A, lane VII). Both the pRG-Dis and the pRG-R2 constructs revealed no expression under promotorless conditions with a basic level under promotor-driven conditions. Low expression levels of RLuc (first cistron) from the promoterless constructs can be explained by the presence of leaky transcription from the vector backbone. In conclusion, these results render cryptic promoter activity as an alternative reason for the regulation of the second cistron unlikely.
FIGURE 3.
Exclusion of cryptic promoter and/or splicing activity in the coding region of Cx43. A, Western blot of pRG Di-cis constructs with (+) or without (−) CMV promoter. NIH3T3 cells were transiently transfected with the control Di-cis vector pRG Di-cis (lanes I and II), pRG-R1 (lanes III and IV), pRG-R2 (lanes V and VI), and pRG-R3 construct (lanes VII and VIII). Western blot analysis of promoterless constructs (−) (lanes II, IV, VI, and VIII) revealed loss of EGFP (second cistron) expression as compared with constructs with CMV promoter (+) (lanes I, III, V, and VII). Faint RLuc expression from these constructs (first cistron) can be explained by the presence of leaky transcription from the vector backbone. Note that the β-actin protein band represents equal loading of protein samples. *, nonspecific labeling of the anti-FLuc antibody. B, outline of PCR strategy of Di-cis constructs pRg-R1, pRg-R2, and pRg-3 to exclude cryptic splice variants. Forward primer D1 (half-arrow) was designed ahead of the chimeric intron, whereas reverse primer D2 (half-arrow) was designed in the coding region of FLuc. C, RT-PCR of Di-cis constructs, pRg-R1, pRg-R2, pRg-R3, and control vector pRg. Total RNA was isolated from transiently transfected N2A cells 48 h post-transfection and reverse transcribed to generate cDNA. No template control (NTC), cDNA from non-transfected N2A cells, and normalization by 18 S rRNA served as controls. Note the successive shortening of amplicons reflecting the size of inserts in the Di-cis construct pRg. The lack of additional amplicons rules out the possibility of cryptic splicing.
To further clarify whether cryptic splice variants are involved in activating the downstream cistron (FLuc) in the dicistronic assay, RT-PCR was performed. The RT-PCR strategy is outlined in Fig. 3B. RT-PCR using primer pairs D1/D2 and the cDNA template from the Di-cis construct R1 (nt 1–637), R2 (nt 1–442), and R3 (nt 442–637) generated single amplicons (Fig. 3C) of ∼1.7 kb (lane 1), ∼1.5 kb (lane 2), and ∼1.26 kb (lane 3), respectively. Similarly, PCR using primer pairs D1/D2 and plasmid DNA as a template from R1, R2, and R3 constructs generated amplicons of ∼1.85 kb (lane 1), ∼1.64 kb (lane 2), and ∼1.4 kb (lane 3). The detection of single amplicons in all Di-cis constructs rules out the possibility of cryptic splicing as an alternative mechanism of separate generation of CT domains of Cx43.
IRES Activity Is Propagated under Hypoxic Conditions
Recent reports have indicated that the expression of an endogenous carboxyl-terminal domain of Cx43 is enhanced under ischemic conditions (4). To investigate a link between ischemic/hypoxic conditions and IRES activity, we transiently transfected NIH3T3 with the Di-cis constructs. 40 h post-transfection, cells were subjected to CIH. After CIH treatment, cell extracts were prepared and run on 12% SDS-PAGE. Immunodetection using RLuc and EGFP antibodies showed that the CIH treatment of pRG-R1 resulted in a detectable increase in EGFP expression (Fig. 4A, lane IV) as compared with untreated pRG-R1 construct (Fig. 4A, lane III). No change in the expression of EGFP was observed from the control pRG-Di-cis (lanes I and II). Similarly, from the pRG-R2 construct (lanes V and VI), no change in the expression of EGFP was observed after CIH treatment. However, CIH treatment of the pRG-R3 construct resulted in an appreciable increase in EGFP expression (Fig. 4A, lane VIII) as compared with the untreated pRG-R3 construct (lane VII). CIH treatment did not affect the expression of the first cistron, as compared with individual untreated constructs (Fig. 4A, RLuc). Quantitative evaluation of the different expression levels is indicated in Fig. 4B. These results suggest that CIH is influential on the activity of the second cistron (EGFP) and hence is likely to be involved in affecting the IRES activity.
FIGURE 4.
CIH increases IRES activity. A, Western blot of pRG Di-cis constructs with (+) or without (−) CIH. NIH3T3 cells were transiently transfected with control pRG Di-cis (lanes I and II), pRG-R1 construct (lanes III and IV), pRG-R2 (lanes V and VI), and pRG-R3 constructs (lanes VII and VIII). CIH treatment of R1 and R3 constructs resulted in an appreciable increase in the expression of EGFP, indicative of increased IRES activity after CIH treatment. In control pRG-Di-cis construct (lanes I and II) and in R2 construct, no appreciable increase in EGFP expression was detectable after CIH treatment. Expression of RLuc of the respective constructs remained constant without (−) or with (+) CIH treatment. The β-actin protein band represents equal loading of protein samples. B, densitometric assessment of EGFP expression of three independent Western blots as shown in A. Data are expressed as mean ± S.E. (error bars) (*, p < 0.05). C, CIH treatment of cultured astrocytes shows a change in the expression pattern of HMBs over time with increasingly dominant expression of the HMB0 isoform. D, quantitative analysis of Western blots of HMBs after CIH. Lower (HMB0) was measured separately from the cumulative evaluation of both upper migrating forms (HMB1 and HMB2). ***, p < 0.001. n = 4 independent experiments.
Regulation of IRES activity under ischemic/hypoxic conditions in the heterologous expression system prompted us to investigate effects of CIH on endogenous Cx43 expression in mouse primary astrocytes. For this purpose, CIH was induced in mouse primary astrocytes for different time points. Immunodetection showed that the CIH treatment resulted in a remarkable change in the expression pattern of HMBs with an overtime increase of the fastest migrating form (HMB0) and concurrent decrease of the middle (HMB1) and the upper band (HMB2) (Fig. 4C). Similar changes in the full-length isoforms of Cx43 after CIH treatment of astrocytes have been described previously (22) with a marked increase of the “dephosphorylated” isoform (P0). From the above data it can be argued that the HMBs comprise different phosphorylated forms of CT-Cx43 fragments. Treatment of lysates from Cx43-transfected NIH3T3 with potato phosphatase corroborates this idea because both upper bands disappear with the persistence of the HMB0 form (Fig. 5B). To further evaluate whether the shift in the HMBs is a sole effect of dephosphorylation of the HMB1 and HMB2 forms and lacks a contribution of de novo synthesis by IRES activity, we performed quantitative Western blot analysis of CIH-treated astrocytes. We reasoned that during the course of CIH treatment, an increase of the total amount of protein (total intensity of all three HMBs) would appear. As shown in Fig. 4D, which indicates expression of the accumulated amounts of HMBs over a period from 0 to 120 min, a significant increase of the proteins became prevalent.
In Vivo Ischemia of Brain Tissue Also Influences the Expression of HMBs
We finally assessed the effect of hypoxia on the expression of HMBs under in situ conditions. For this purpose, we used an animal model (24) that allows a reproducible and controlled hypoxia-ischemia in young rats (25). Animals with unilateral ligature of the common carotid artery and subsequent systemic hypoxia for 90 min were sampled and processed for Western blot analysis. Hemispheres of animals without hypoxic/ischemic treatment served as controls. Control animals revealed full-length bands around 43 kDa and HMBs in the form of a doublet band around 27 kDa and a single band at 20 kDa (note that in this case the proteins were loaded on a 15% SDS gel, which leads to a change in the separation pattern of the fast migrating bands (Fig. 5A); see “Experimental Procedures”). Induction of ischemia/hypoxia led to a significant increase of the higher mobility bands with a dominance of the HMB0 band (Fig. 5A). Comparison of the total amount of HMBs (Fig. 5C) clearly indicates a significant increase (p < 0.01) in favor of de novo synthesis, thereby confirming the in vitro experiments with CIH-treated astrocytes. Equal loading was controlled by β-actin controls to confirm equal sample loading (Fig. 5B). We conclude that the in situ data provide a further compelling argument in favor of a dynamic expression of high mobility fragments of Cx43 and its modifiability by hypoxic conditions.
LC-MC Identifies HMB0 as a CT-Cx43 Fragment
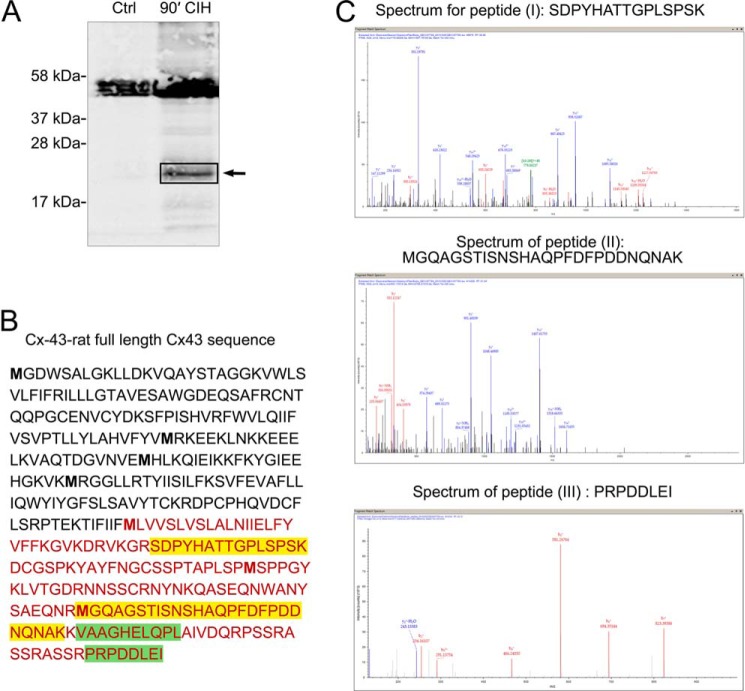
To finally prove the identity of the HMBs, we performed mass spectrometry (LC-MS/MS) on primary rat astrocytes. The samples were taken from ischemic treated astrocytes (CIH; see above) to obtain sufficient amounts of HMB0 protein. Lysates were transferred to SDS-PAGE analysis. A first gel was counterstained with Coomassie Brilliant Blue for subsequent in-gel digestion with trypsin, whereas a parallel gel was used for proper identification of HMB0 by Western blotting with Cx43 antibodies (Fig. 6A).
FIGURE 6.
Mass spectrometry of HMB0 slices identifies carboxyl-terminal peptides of Cx43. A, Western blot of a gel used for MS analysis. Cultures of neonatal rat astrocytes, which underwent CIH treatment for 90 min, show enhanced HMB0 staining with anti-Cx43 antibodies (90′, CIH) in comparison with untreated controls (Ctrl). Gel bands of the corresponding HMB0 height were cut out (see arrow) and subjected to mass spectrometry. B, nano-HPLC and electrospray ionization-MS/MS spectrometry after tryptic digestion identified four different peptides, which can be assigned to the carboxyl terminus of Cx43. Two peptides (labeled I and II and indicated in yellow) were achieved at highest stringency (search for miscleavage set to 1, meaning no miscleavage). Peptide III (indicated in green) was achieved when miscleavage search criteria were set to 2 (meaning one miscleavage). Peptide IV (VAAGHELQPL) (also highlighted in yellow) was achieved with a “no enzyme” search. A nonspecific cleavage event could have generated this fragment during processing of the sample. Alternatively, the fragment represents an in vivo proteolytically processed fragment, which might have co-migrated with intact HMB0. Note that no further sequence was identified outside the carboxyl terminus. C, corresponding MS/MS spectra (b and y fragmentation series) of peptides I, II, and III. All raw data in the form of an msf file are posted under QEXII07709.msf.
Mass spectrometry data obtained from the digest of cut-out HMB0 gel stripes identified at least two fully tryptic peptides (Fig. 6A) that fulfill the conventional criteria for annotating them as carboxyl-terminal fragments of Cx43. Two further peptides, which were obtained at lower stringency search conditions, also comprise domains of the C terminus of Cx43; their definite assignment, however, remains disputable (see the legend to Fig. 6, B and C).
DISCUSSION
Immunoblots of Cx43 from various tissues and cells are known to detect some low molecular weight protein band(s), which are regarded as carboxyl-terminal domains (4, 17). Usually two to three higher mobility bands (HMBs) are detected with antibodies directed against the carboxyl-terminal domain of Cx43. The existence of one of these protein bands was found to depend on the phosphorylated state of Cx43. When the full-length Cx43 occurs in the unphosphorylated isoform, only one higher mobility band is found to exist (17). Our own data substantiate these findings by showing a concurrent increase of the HMB0 form under hypoxic conditions, when the P1 and P2 isoforms of full-length Cx43 decreases (22). Apart from this change in phosphorylation pattern after hypoxic treatment, an increase of the total amount of HMBs was apparent, as indicated by quantitative Western blotting.
To explore the possible molecular mechanism for the generation of separate CT-Cx43 fragments, we investigated whether protease cleavage is responsible for the occurrence of CT-Cx43 domains. This mechanism was ruled out using frameshift experiments that excluded the expression of full-length Cx43. Introduction of a T nucleotide at position 590 led to a complete loss of full-length protein with persisting HMBs. A downstream mutation (nt 712) corrupted the expression of both translational products (full-length and HMBs). To investigate the internal translation of this carboxyl-terminal fragment by an internal IRES element, we subcloned different DNA fragments (from the coding region of Cx43) in a dicistronic vector. Dicistronic assays indicated that a DNA sequence (nt 442–637), which is located immediately upstream of the CT-Cx43 coding region, possesses IRES activity. This putative IRES activity was subjected to stringent controls to exclude the presence of cryptic promoters or cryptic splicing events as alternative reasons for the generation of CT-Cx43 fragments (26).
The existence of IRES elements in the coding region of eukaryotic genes has been reported in previous studies (27–29). In a recent report, we provided first evidence of an internal IRES-mediated translation of a carboxyl-terminal fragment in the connexin gene family (30). In this case, zebrafish connexin zfCx55.5 revealed an alternate transcript of about 11 kDa of the carboxyl terminus (p11-CT), which was found to translocate to the nucleus in heterologous expression systems. A concurrent study (31) yielded insight into the molecular mechanism linking IRES activity in the zfCx55.5 gene to binding of polypyrimidine tract-binding protein, an important IRES trans-acting factor. This finding is noteworthy because polypyrimidine tract-binding protein appears to serve a dual function in splicing and IRES-mediated translation (32). Unlike in our previous work, we were unable to identify a putative AUG codon from which the internal translation of CT-Cx43 starts. Deletion of an AUG codon, present in a favorable position (nt 638 in the coding region) for initiation of CT-Cx43 translation, did not interfere with its expression. Extending the deletion from nt 638 to 714, however, resulted in complete loss of the alternate transcript. This observation was supported by frameshift experiments, which delineate a region between nt 638 and 714 as important for the expression of HMBs. A feasible explanation for a possible lack of a canonical initiation codon derives from observations that showed that translation initiation can occur from any codon and, more importantly, without the participation of initiator methionine tRNA (33). Additionally, IRES elements are known to exhibit specific secondary structures, which seemingly play crucial roles in determining their activity (34, 35) and thus may be important for the selection of initiation codons. It needs to be further explored whether a similar mechanism plays a role in the initiation of translation of the CT-Cx43 fragment. It is noteworthy that an internal ribosomal entry site for Cx43 has been reported, residing in the 5′-UTR of Cx43 (36). Regulation of its activity, including a further connexin (Cx26) mRNA was described under CAP-independent density-inhibited regulation of cell proliferation in human pancreatic tumor cells (37). Although speculative at our current state of knowledge, a differential regulation of both IRES activities by means of separate IRES trans-acting factors seems a feasible option (38). One would expect that different functions of Cx43 are requested, depending on the initial biological challenge, which necessitates the full-length version for channel formation or the carboxyl-terminal domain for non-channel functions.
Separate expression of the HMB0 domain, which we finally identified by mass spectrometry as a carboxyl-terminal segment, is significant in terms of Cx43 functions. In fact, Cx43 and/or CT-Cx43 domains have been suggested to influence important biological activities like regulation of cell growth (39) and resistance to cell death (40) by mechanisms that do not require gap junction and/or hemichannel communication (13, 14). These channel-independent functions necessitate a cytoplasm to nucleus signaling either by cytoplasmic generated messengers (i.e. growth regulators) (41, 42) or by a direct role of CT-Cx43 domains. A separate expression of biologically active domains and its nuclear translocation may endow Cx43 proteins with the capability of modulating gene expression related to growth control and cell survival. The endogenously produced HMBs may thus represent molecular determinants for a direct regulatory link between the cytoplasm and nucleus, as has been suggested in previous works (4, 15).
The activity of IRES elements has been shown to respond to stress conditions, like γ-radiation (43), hypoxia (44), or amino acid starvation (45). Consistent with this concept, we found that the activity of the putative IRES element present in the coding region of Cx43 was enhanced during chemically induced ischemic deprivation (CIH) in astrocytes and in ischemic brains. This result corroborates observations that expression of endogenously CT-Cx43 domains is significantly increased under hypoxic conditions in cardiomyocytes and after global ischemia of the isolated perfused rat heart (4). It is also in accordance with recent observations on transgenic mice carrying a transgene with a carboxyl-terminal truncated Cx43. Ablation of the Cx43-carboxyl terminus was shown to increase susceptibility to cell death and secondary ischemic events, such as invasion of inflammatory cells (10). Notably, our data indicate that hypoxic/ischemic treatment increased IRES activity and may influence the phosphorylation state of full-length Cx43 and its HMBs.
From the above, we conclude that metabolic conditions are apt to influence expression of CT-Cx43 fragments that endow the Cx43 gene with an independent regulatory role in cell metabolism and cell survival, a challenging perspective that requires further intensive experimental exploration.
Addendum
During the final preparation of our revised manuscript, Smyth and Shaw published a paper (46) where they report on the same subject. Although the experimental approaches are different, both papers come to the same conclusions.
This work was supported by the International Graduate School of Neuroscience of NRW/Germany (to M. H. and S. O.), German Research Council (Deutsche Forschungsgemeinschaft (DFG), Graduate College 736, Development and Plasticity of the Nervous System) (to B. W.), and DFG Grant SFB 509 (to R. D. and G. Z.).
- HMB
- higher mobility band
- CIH
- chronic intermittent hypoxia
- CT
- carboxyl terminus
- nt
- nucleotide(s)
- EGFP
- enhanced GFP
- IRES
- internal ribosomal entry site
- FLuc
- firefly luciferase
- RLuc
- Renilla luciferase
- Di-cis
- dicistronic.
REFERENCES
- 1. Willecke K., Eiberger J., Degen J., Eckardt D., Romualdi A., Güldenagel M., Deutsch U., Söhl G. (2002) Structural and functional diversity of connexin genes in the mouse and human genome. Biol. Chem. 383, 725–737 [DOI] [PubMed] [Google Scholar]
- 2. Evans W. H., Martin P. E. (2002) Gap junctions: structure and function. Mol. Membr. Biol. 19, 121–136 [DOI] [PubMed] [Google Scholar]
- 3. Márquez-Rosado L., Solan J. L., Dunn C. A., Norris R. P., Lampe P. D. (2012) Connexin43 phosphorylation in brain, cardiac, endothelial and epithelial tissues. Biochim. Biophys. Acta 1818, 1985–1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kardami E., Dang X., Iacobas D. A., Nickel B. E., Jeyaraman M., Srisakuldee W., Makazan J., Tanguy S., Spray D. C. (2007) The role of connexins in controlling cell growth and gene expression. Prog. Biophys. Mol. Biol. 94, 245–264 [DOI] [PubMed] [Google Scholar]
- 5. Omori Y., Yamasaki H. (1998) Mutated connexin43 proteins inhibit rat glioma cell growth suppression mediated by wild-type connexin43 in a dominant-negative manner. Int. J. Cancer 78, 446–453 [DOI] [PubMed] [Google Scholar]
- 6. Santiago M. F., Alcami P., Striedinger K. M., Spray D. C., Scemes E. (2010) The carboxyl-terminal domain of connexin43 is a negative modulator of neuronal differentiation. J. Biol. Chem. 285, 11836–11845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Elias L. A., Wang D. D., Kriegstein A. R. (2007) Gap junction adhesion is necessary for radial migration in the neocortex. Nature 448, 901–907 [DOI] [PubMed] [Google Scholar]
- 8. Crespin S., Bechberger J., Mesnil M., Naus C. C., Sin W. C. (2010) The carboxy-terminal tail of connexin43 gap junction protein is sufficient to mediate cytoskeleton changes in human glioma cells. J. Cell Biochem. 110, 589–597 [DOI] [PubMed] [Google Scholar]
- 9. Cina C., Maass K., Theis M., Willecke K., Bechberger J. F., Naus C. C. (2009) Involvement of the cytoplasmic C-terminal domain of connexin43 in neuronal migration. J. Neurosci. 29, 2009–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kozoriz M. G., Bechberger J. F., Bechberger G. R., Suen M. W., Moreno A. P., Maass K., Willecke K., Naus C. C. (2010) The connexin43 C-terminal region mediates neuroprotection during stroke. J. Neuropathol. Exp. Neurol. 69, 196–206 [DOI] [PubMed] [Google Scholar]
- 11. King T. J., Bertram J. S. (2005) Connexins as targets for cancer chemoprevention and chemotherapy. Biochim. Biophys. Acta 1719, 146–160 [DOI] [PubMed] [Google Scholar]
- 12. Mesnil M., Crespin S., Avanzo J. L., Zaidan-Dagli M. L. (2005) Defective gap junctional intercellular communication in the carcinogenic process. Biochim. Biophys. Acta 1719, 125–145 [DOI] [PubMed] [Google Scholar]
- 13. Zhang Y. W., Kaneda M., Morita I. (2003) The gap junction-independent tumor-suppressing effect of connexin 43. J. Biol. Chem. 278, 44852–44856 [DOI] [PubMed] [Google Scholar]
- 14. Moorby C., Patel M. (2001) Dual functions for connexins: Cx43 regulates growth independently of gap junction formation. Exp. Cell Res. 271, 238–248 [DOI] [PubMed] [Google Scholar]
- 15. Dang X., Doble B. W., Kardami E. (2003) The carboxy-tail of connexin-43 localizes to the nucleus and inhibits cell growth. Mol. Cell Biochem. 242, 35–38 [PubMed] [Google Scholar]
- 16. Laird D. W., Puranam K. L., Revel J. P. (1991) Turnover and phosphorylation dynamics of connexin43 gap junction protein in cultured cardiac myocytes. Biochem. J. 273, 67–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Joshi-Mukherjee R., Coombs W., Burrer C., de Mora I. A., Delmar M., Taffet S. (2007) Evidence for the presence of a free C-terminal fragment of cx43 in cultured cells. Cell Commun. Adhes. 14, 75–84 [DOI] [PubMed] [Google Scholar]
- 18. Hudder A., Werner R. (2000) Analysis of a Charcot-Marie-Tooth disease mutation reveals an essential internal ribosome entry site element in the connexin-32 gene. J. Biol. Chem. 275, 34586–34591 [DOI] [PubMed] [Google Scholar]
- 19. Theis M., Jauch R., Zhuo L., Speidel D., Wallraff A., Döring B., Frisch C., Söhl G., Teubner B., Euwens C., Huston J., Steinhäuser C., Messing A., Heinemann U., Willecke K. (2003) Accelerated hippocampal spreading depression and enhanced locomotory in mice with astrocyte-directed inactivation of connexin43. J. Neurosci. 23, 766–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Teubner B., Michel V., Pesch J., Lautermann J., Cohen-Salmon M., Söhl G., Jahnke K., Winterhager E., Herberhold C., Hardelin J. P., Petit C., Willecke K. (2003) Connexin30 (Gjb)-deficiency causes severe hearing impairment and lack of endocochlear potential. Hum. Mol. Genet. 12, 13–21 [DOI] [PubMed] [Google Scholar]
- 21. Dermietzel R., Hertberg E. L., Kessler J. A., Spray D. C. (1991) Gap junctions between cultured astrocytes: immunocytochemical, molecular, and electrophysiological analysis. J. Neurosci. 11, 1421–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li W., Hertzberg E. L., Spray D. C. (2005) Regulation of connexin43-protein binding in astrocytes in response to chemical ischemia/hypoxia. J. Biol. Chem. 280, 7941–7948 [DOI] [PubMed] [Google Scholar]
- 23. Contreras J. E., Sánchez H. A., Eugenin E. A., Speidel D., Theis M., Willecke K., Bukauskas F. F., Bennett M. V., Sáez J. C. (2002) Metabolic inhibition induces opening of unapposed connexin 43 gap junction hemichannels and reduces gap junctional communication in cortical astrocytes in culture. Proc. Natl. Acad. Sci. U.S.A. 99, 495–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Levine S., Klein M. (1960) Ischemic infarction and swelling in the rat brain. Arch. Pathol. 69, 544–553 [PubMed] [Google Scholar]
- 25. Meier C., Middelanis J., Wasielewski B., Neuhoff S., Roth-Haerer A., Gantert M., Dinse H. R., Dermietzel R., Jensen A. (2006) Spastic paresis after perinatal brain damage in rats is reduced by human cord blood mononuclear cells. Pediatr. Res. 59, 244–249 [DOI] [PubMed] [Google Scholar]
- 26. Van Eden M. E., Byrd M. P., Sherrill K. W., Lloyd R. E. (2004) Demonstrating internal ribosome entry sites in eukaryotic mRNAs using stringent RNA test procedures. RNA 10, 720–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cornelis S., Bruynooghe Y., Denecker G., Van Huffel S., Tinton S., Beyaert R. (2000) Identification and characterization of a novel cell cycle-regulated internal ribosome entry site. Mol. Cell 5, 597–605 [DOI] [PubMed] [Google Scholar]
- 28. Lauring A. S., Overbaugh J. (2000) Evidence that an IRES within the Notch2 coding region can direct expression of a nuclear form of the protein. Mol. Cell 6, 939–945 [DOI] [PubMed] [Google Scholar]
- 29. Herbreteau C. H., Weill L., Décimo D., Prévôt D., Darlix J. L., Sargueil B., Ohlmann T. (2005) HIV-2 genomic RNA contains a novel type of IRES located downstream of its initiation codon. Nat. Struct. Mol. Biol. 12, 1001–1007 [DOI] [PubMed] [Google Scholar]
- 30. Ul-Hussain M., Zoidl G., Klooster J., Kamermans M., Dermietzel R. (2008) IRES-mediated translation of the carboxy-terminal domain of the horizontal cell specific connexin Cx55.5 in vivo and in vitro. BMC Mol. Biol. 9, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ul-Hussain M., Dermietzel R., Zoidl G. (2008) Characterization of the internal IRES element of the zebrafish connexin55.5 reveals functional implication of the polypyrimidine tract binding protein. BMC Mol. Biol. 9, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ul-Hussain M., Dermietzel R., Zoidl G. (2012) Connexins and Cap-independent translation: role of internal ribosome entry sites. Brain Res. 148, 99–106 [DOI] [PubMed] [Google Scholar]
- 33. RajBhandary U. L. (2000) More surprises in translation: initiation without the initiator tRNA. Proc. Natl. Acad. Sci. U.S.A. 97, 1325–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sasaki J., Nakashima N. (2000) Methionine-independent initiation of translation in the capsid protein of an insect RNA virus. Proc. Natl. Acad. Sci. U.S.A. 97, 1512–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kanamori Y., Nakashima N. (2001) A tertiary structure model of the internal ribosome entry site (IRES) for methionine-independent initiation of translation. RNA 7, 266–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schiavi A., Hudder A., Werner R. (1999) Connexin43 mRNA contains a functional internal ribosome entry site. FEBS Lett. 464, 118–122 [DOI] [PubMed] [Google Scholar]
- 37. Lahlou H., Fanjul M., Pradayrol L., Susini C., Pyronnet S. (2005) Restoration of functional gap junctions through internal ribosome entry site-dependent synthesis of endogenous connexins in density-inhibited cancer cells. Mol. Cell Biol. 25, 4034–4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vagner S., Galy B., Pyronnet S. (2001) Irresistible IRES. Attracting the translation machinery to internal ribosome entry sites. EMBO Rep. 2, 893–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Olbina G., Eckhart W. (2003) Mutations in the second extracellular region of connexin 43 prevent localization to the plasma membrane, but do not affect its ability to suppress cell growth. Mol. Cancer Res. 1, 690–700 [PubMed] [Google Scholar]
- 40. Cusato K., Bosco A., Rozental R., Guimarães C. A., Reese B. E., Linden R., Spray D. C. (2003) Gap junctions mediate bystander cell death in developing retina. J. Neurosci. 23, 6413–6422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gellhaus A., Dong X., Propson S., Maass K., Klein-Hitpass L., Kibschull M., Traub O., Willecke K., Perbal B., Lye S. J., Winterhager E. (2004) Connexin43 interacts with NOV: a possible mechanism for negative regulation of cell growth in choriocarcinoma cells. J. Biol. Chem. 279, 36931–36942 [DOI] [PubMed] [Google Scholar]
- 42. Fu C. T., Bechberger J. F., Ozog M. A., Perbal B., Naus C. C. (2004) CCN3 (NOV) interacts with connexin43 in C6 glioma cells: possible mechanism of connexin-mediated growth suppression. J. Biol. Chem. 279, 36943–36950 [DOI] [PubMed] [Google Scholar]
- 43. Holcik M., Lefebvre C., Yeh C., Chow T., Korneluk R. G. (1999) A new internal-ribosome-entry-site motif potentiates XIAP-mediated cytoprotection. Nat. Cell Biol. 1, 190–192 [DOI] [PubMed] [Google Scholar]
- 44. Stein I., Itin A., Einat P., Skaliter R., Grossman Z., Keshet E. (1998) Translation of vascular endothelial growth factor mRNA by internal ribosome entry: implications for translation under hypoxia. Mol. Cell Biol. 18, 3112–3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fernandez J., Yaman I., Mishra R., Merrick W. C., Snider M. D., Lamers W. H., Hatzoglou M. (2001) Internal ribosome entry site-mediated translation of a mammalian mRNA is regulated by amino acid availability. J. Biol. Chem. 276, 12285–12291 [DOI] [PubMed] [Google Scholar]
- 46. Smyth J. W., Shaw R. M. (2013) Autoregulation of connexin43 gap junction formation by internally translated isoforms. Cell Rep. 5, 611–618 [DOI] [PMC free article] [PubMed] [Google Scholar]