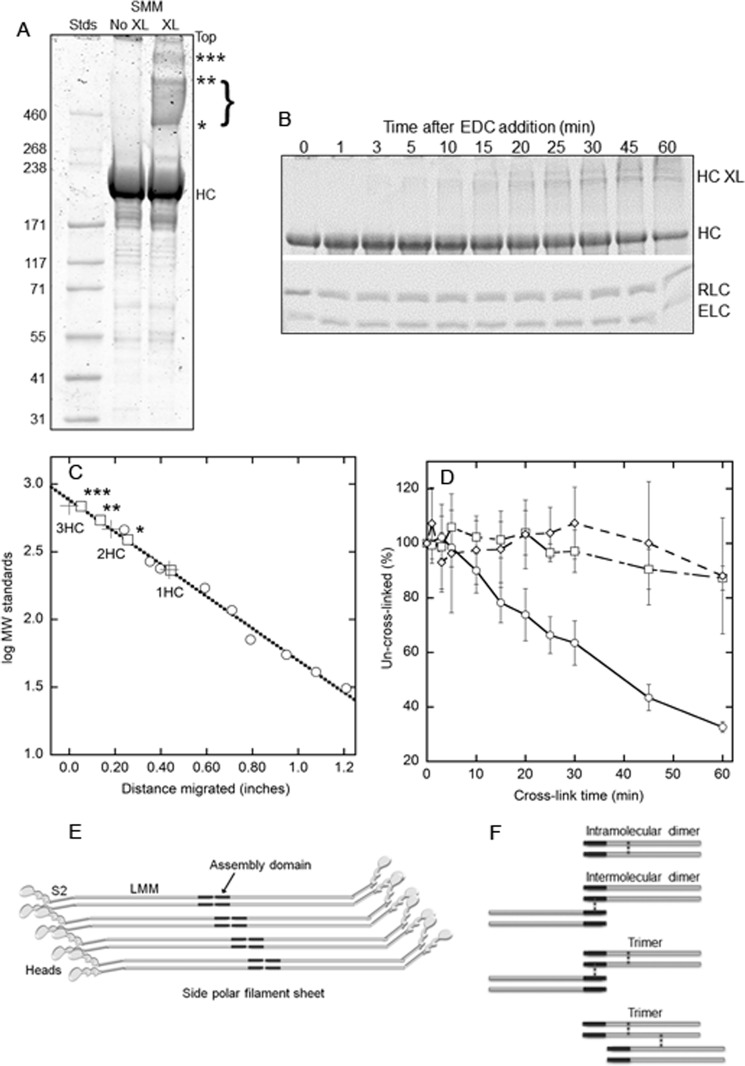
FIGURE 1.
EDC cross-linking of Rh-SMM filaments. A, image of Coomassie-stained 3–8% Tris-acetate gel (Invitrogen) showing HiMark prestained high molecular weight protein standards (Stds; Invitrogen), 5 μg of Rh-SMM (No XL), and 5 μg of XL-Rh-SMM (XL; 30-min EDC reaction). Less than 5% of the protein in this sample remained in the supernatant after pelleting the filaments. Un-cross-linked HC, cross-linked heavy chain bands are labeled with asterisks. Light chains are not seen on this gel. B, split image of one 4–20% Tris-glycine gel (Invitrogen) showing time course of EDC cross-linking of Rh-SMM filaments quenched with 5 mm DTT at specified times. Upper panel, bands labeled HC XL correspond to those from A, although they are not well resolved; lower panel, light chain region. C, plot of molecular mass standards (circles) fit to a line (dotted). Crosses are the calculated masses for one, two, or three HC as indicated. Squares show measured band positions corresponding to gel in A, using the standard curve. D, plot of pixel density of bands averaged from eight different EDC cross-linking time courses from three independent Rh-SMM preparations. HC (circles), RLC (squares), and essential light chain (ELC, diamonds). Error bars are ± S.D. E, cartoon showing mode of SMM molecule packing in a single side polar filament sheet. The tails are shown as gray rods with the assembly domains in black. The S2 region is connected to the light meromyosin (LMM) region of the rod at a known bending region (32). Filament stability comes from intermolecular interactions between assembly domains. F, cartoon of possible types of inter- and intramolecular cross-links induced by EDC treatment. The positions of the cross-links (dotted vertical lines) are shown in the LMM region of the heavy chains for illustration only. Actual cross-link locations are unknown except that they only involve the heavy chains and that some cross-links must be forming between the assembly domains of adjacent molecules to explain filament stability.