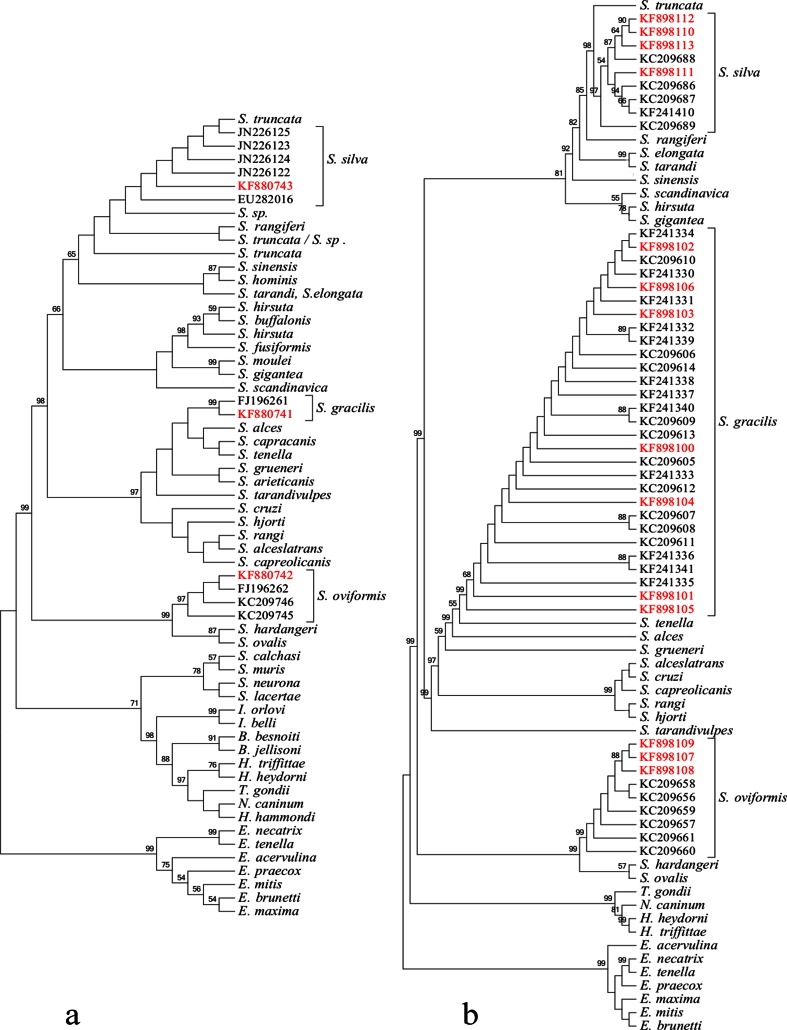
Fig. 2.
Maximum parsimony of DNA phylograms of selected Sarcocystidae and Coccidia. a The ssu rRNA tree was constructed based on the alignment of nearly complete ssu rRNA gene sequences of 14 Polish Sarcocystis spp. isolates and available ssu rRNA gene sequences of related species deposited in GenBank. b The cox1 tree was constructed based on the alignment of nearly complete ssu rRNA gene sequences of 14 Polish Sarcocystis spp. isolates and available ssu rRNA gene sequences of related species deposited in GenBank. Trees were rooted with Eimeria spp. The italicized values among the branches indicate percent bootstrap value per 1,000 replicates. Bootstrap values below 50 % are not shown. Polish isolates are marked in red. The GenBank accession numbers of all sequences used for construction of the trees are given in Table S1 (color figure online)