Figure 3.
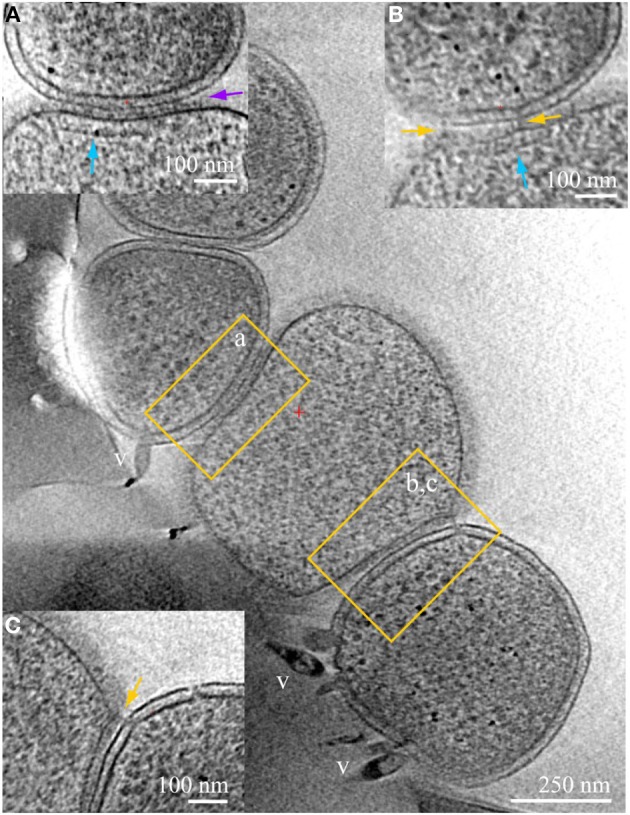
ARMAN—Thermoplasmatales inter-species associations and viral infection processes. Slices from a 3D cryo-ET reconstruction of a colony of cells showing contacts and associations between a Thermoplasmatal lineage cell (center, no cell wall) and two ARMAN cells infected with viruses (gram-negative like cell wall, top left, and bottom right). Insets represent slices at different orientations, of the regions within yellow boxes in the main z-slice. In inset (A) the cell surfaces are in tight contact, with a distinct line of density at the interface pointed by the purple arrow. Thermoplasma lineage archaea cells engaged in this type of association typically have a cytoplasmic band of density parallel to the plasma membrane, indicated here by the vertical blue arrow. These features resemble, to some extent, bacterial chemotaxis apparatus (Milne and Subramaniam, 2009), but AMD Thermoplasmatales lineage archaea have no identified chemotaxis genes. Inset (B) is a slice at an orientation that exactly contains a filamentous connection between the cells that clearly crosses the cell wall; the same band of density as in (B) is indicated by a blue arrow. Inset (C) is another slice of the same region, at different coordinates (z-height and angles), with pili-like connections through the cell wall. The ARMAN cells involved in the interactions shown in insets (A–C) are infected by rod-shape and lemon shaped viruses some of which are clear and labeled “v.” See also Figures S2–S6 and Movie 4.
