Abstract
The southern plains woodrat (Neotoma micropus) is the principal host of Catarina virus in southern Texas and a natural host of other North American Tacaribe serocomplex viruses. The objectives of this study were to increase our knowledge of the genetic diversity among Tacaribe serocomplex viruses associated with N. micropus and to define better the natural host relationships of these viruses. Pairwise comparisons of complete glycoprotein precursor gene sequences and complete nucleocapsid protein gene sequences revealed a high level of genetic diversity among Tacaribe serocomplex viruses associated with N. micropus in western Oklahoma, southern New Mexico, and northern and southern Texas. Collectively, the results of Bayesian analyses of nucleotide sequences and pairwise comparisons of amino acid sequences confirmed that the arenaviruses associated with N. micropus in Oklahoma and New Mexico should be included in the Whitewater Arroyo species complex, and indicated that that the arenaviruses associated with N. micropus in northern Texas are strains of a novel arenaviral species – tentatively named “Middle Pease River virus”. Together, the results of assays for arenavirus and assays for anti-arenavirus antibody in 54 southern plains woodrats and 325 other rodents captured at 2 localities suggested that the southern plains woodrat is the principal host of Middle Pease River virus in northern Texas.
1. Introduction
The North American members of the Tacaribe serocomplex (family Arenaviridae, genus Arenavirus) include Bear Canyon virus (BCNV), Big Brushy Tank virus (BBTV), Catarina virus (CTNV), Real de Catorce virus (RCTV), Skinner Tank virus (SKTV), Tamiami virus (TAMV), Tonto Creek virus (TTCV), and Whitewater Arroyo virus (WWAV) (Cajimat et al. 2011). The results of a previous study (Milazzo et al. 2011) suggested that WWAV or Tacaribe serocomplex viruses antigenically closely related to WWAV are etiological agents of severe febrile illnesses in humans in the United States.
Specific members of the rodent family Cricetidae (Musser and Carleton 2005) are the principal hosts of the Tacaribe serocomplex viruses for which natural host relationships have been well characterized. For example, the southern plains woodrat (Neotoma micropus) in southern Texas is the principal host of CTNV (Milazzo et al. 2013).
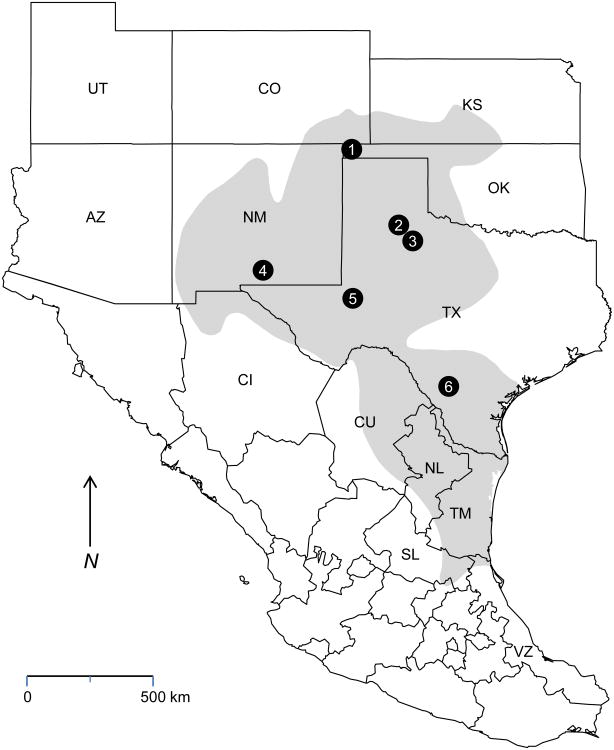
The geographical range of N. micropus extends from southern Kansas and western New Mexico through San Luis Potosí (Figure 1). Previous studies revealed a high level of genetic diversity among Tacaribe serocomplex viruses associated with N. albigula in Arizona and New Mexico (Milazzo et al. 2008); a high level of genetic diversity among Tacaribe serocomplex viruses associated with N. mexicana in Arizona, Colorado, New Mexico, and Utah (Cajimat et al. 2008); and evidence of arenaviral infections in southern plains woodrats captured in Colorado, New Mexico, Oklahoma, and Texas (Cajimat et al. 2011, Calisher et al. 2001, Fulhorst et al. 2002, Milazzo et al. 2010). The purpose of this study was to extend our knowledge of the genetic diversity among Tacaribe serocomplex viruses associated with N. micropus and to define better the natural host relationships of these viruses.
Figure 1.
Map showing the geographical range of Neotoma micropus (shaded area), The localities at which the southern plains woodrats in this study were captured are marked with filled circles: 1 – Black Mesa State Park and Nature Preserve, Cimarron County, Oklahoma; 2 – privately owned property near the town of Flomot and along the Middle Pease River in Motley County, Texas; 3 – privately owned property near the town of Afton, Dickens County, Texas; 4– Fort Bliss Military Base in Otero County, New Mexico; 5 – Monahans Sandhills State Park, Ward County, Texas; 6 – Chaparral Wildlife Management Area, Dimmit and La Salle counties, Texas. The properties in Motley and Dickens counties were separated by 53.5 km. United States – AZ, Arizona; CO, Colorado; KS, Kansas; NM, New Mexico; OK, Oklahoma; TX, Texas; UT, Utah. Mexico – CI, Chihuahua; CU, Coahuila; NL, Nuevo León; QU, Querétaro; SL, San Luis Potosí; TM, Tamaulipas; VZ, Veracruz. Map adapted from Patterson et al. 2007.
2. Materials and Methods
The rodents in this study were from Black Mesa State Park and Nature Preserve (BMSP) in Cimarron County, Oklahoma; Fort Bliss Military Base (FBMB) in Otero County, New Mexico; a 160-ha cattle ranch located 1.6 km south of the town of Flomot (FLOM) in Motley County, Texas; a 350-ha cattle ranch located 1.3 km north of the town of Afton (AFTN) in Dickens County, Texas; Monahans Sandhills State Park (MSSP) in Ward County, Texas; and the Chaparral Wildlife Management Area (CWMA) in Dimmit and La Salle counties, Texas. Previously, arenavirus AV 98490013 was isolated from a southern plains woodrat captured at BMSP (Cajimat et al. 2011); anti-arenavirus antibody was found in southern plains woodrat TK77260, 5 (5.8%) of 86 other rodents captured at FBMB, and 16 (4.1%) of 390 rodents captured at FLOM, AFTN, and MSSP (Table 1); arenavirus AV H0380005 was isolated from woodrat TK77260 (Cajimat et al. 2011); and the 6 CTNV strains in this study (Table 2) were isolated from southern plains woodrats captured at the CWMA in 1999--2004 (Fulhorst et al. 2002, Milazzo et al. 2013). In this study, fresh-frozen samples of kidney and spleen from the rodents captured at FBMB, FLOM, AFTN, and MSSP were acquired from the Museum of Texas Tech University and tested for arenavirus.
Table 1.
Prevalence of anti-arenavirus antibody in rodents captured in New Mexico or Texas, by species and county.a
| Localityb | |||||
|---|---|---|---|---|---|
|
|
|||||
| Species | FBMB | FLOM | AFTN | MSSP | Total |
| Baiomys taylori | -- | 0/10 | 0/5 | -- | 0/15 |
| Neotoma albigula | 0/21 | -- | -- | -- | 0/21 |
| Neotoma micropus | 6/9 | 5/14 | 7/40 | 2/2 | 20/65 |
| Onychomys arenicola | 0/5 | -- | -- | -- | 0/5 |
| Onychomys leucogaster | 0/2 | -- | 0/7 | 0/3 | 0/12 |
| Peromyscus attwateri | -- | 0/3 | 0/15 | -- | 0/18 |
| Peromyscus eremicus | 0/10 | -- | -- | -- | 0/10 |
| Peromyscus leucopus | 0/12 | 0/32 | 0/98 | 0/2 | 0/144 |
| Peromyscus levipes | -- | 0/1 | -- | -- | 0/1 |
| Peromyscus maniculatus | 0/10 | 0/2 | 0/1 | 0/4 | 0/17 |
| Reithrodontomys fulvescens | -- | 0/5 | 0/15 | -- | 0/20 |
| Reithrodontomys megalotis | 0/11 | 0/5 | 0/7 | -- | 0/23 |
| Reithrodontomys montanus | -- | 0/6 | 0/4 | -- | 0/10 |
| Sigmodon hispidus | 0/7 | 0/39 | 2/70 | -- | 2/116 |
| Total | 6/87 | 5/117 | 9/262 | 2/11 | 22/477 |
The antibody data are from Milazzo et al. (2010) and Mauldin et al. (2013).
FBMB, Fort Bliss Military Base in Otero County, New Mexico (1,300 trap nights in 13 nights in May 1998-Oct 1998); FLOM, a ranch near the town of Flomot, Motley County, Texas (2,150 trap-nights in 11 nights, Oct 2006-Oct 2009); AFTN, a ranch near the town of Afton, Dickens County, Texas (1,444 trap-nights in 4 nights, Jun 2008-Apr 2009); MSSP, Monahans Sandhills State Park, Ward County, Texas (500 trap-nights in 5 nights, Jun 2002-Jul 2002).
Table 2.
Arenaviruses naturally associated with Neotoma micropus and included in the analyses of glycoprotein precursor (GPC) gene sequences and nucleocapsid (N) protein gene sequences, by county.
| Woodrat | |||||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|||||||||
| Virusa | Strain | GPC geneb,c | N protein geneb,c | Voucherd | Statee | Countyf | Geographical coordinates | Date captured | Cytb geneg |
| WWAV | AV H0380005 | EU910959 | EU910959 | TK77260 | NM | Otero | 32°27′22″ N, 105°41′04″ W | 5/21/1998 | AF376473 |
| WWAV | AV H0380011 | JX657688 | JX560800 | TK77270 | NM | Otero | 32°28′16″ N, 105°42′05″ W | 5/22/1998 | AF376474 |
| WWAV | AV H0380014 | JX657689 | JX560801 | TK77278 | NM | Otero | 32°31′27″ N, 105°53′04″ W | 5/23/1998 | KC153472 |
| WWAV | AV H0380016 | JX657690 | JX560802 | TK74993 | NM | Otero | 32°28′16″ N, 105°42′05″ W | 5/20/1998 | KC153473 |
| WWAV | AV H0380020 | JX657691 | JX560803 | TK74995 | NM | Otero | 32°27′30″ N, 105°40′58″ W | 5/20/1998 | KC153474 |
| WWAV | AV 98490013 | FJ032026 | FJ032027 | TK28731 | OK | Cimarron | 36°52′50″ N, 102°55′28″ W | 10/12/1985 | FJ716217 |
| MPRV | AV I0130002 | JX657692 | JX560804 | TK137078 | TX | Motley | 34°12′57″ N, 100°59′46″ W | 10/7/2006 | KC153476 |
| MPRV | AV I0130006 | JX560798 | JX560798 | TK137081 | TX | Motley | 34°12′57″ N, 100°59′46″ W | 10/7/2006 | KC153477 |
| MPRV | AV N0030003 | JX657693 | JX560805 | TK160340 | TX | Motley | 34°12′57″ N, 100°59′46″ W | 10/4/2008 | KC153478 |
| MPRV | AV N0030006 | JX657694 | JX560806 | TK166046 | TX | Motley | 34°12′57″ N, 100°59′46″ W | 10/17/2009 | KC153479 |
| MPRV | AV M0040017 | JX560799 | JX560799 | TK147378 | TX | Dickens | 33°45′36″ N, 100°48′44″ W | 9/28/2008 | KC153480 |
| MPRV | AV M0040020 | JX657695 | JX560807 | TK147396 | TX | Dickens | 33°45′32″ N, 100°47′42″ W | 9/28/2008 | KC153481 |
| MPRV | AV M0040023 | JX657696 | JX560808 | TK147425 | TX | Dickens | 33°45′36″ N, 100°48′44″ W | 2/14/2009 | KC153482 |
| MPRV | AV J0220060 | JX657699 | JX560811 | TK147327 | TX | Dickens | 33°45′36″ N, 100°48′44″ W | 6/28/2008 | KC153483 |
| MPRV | AV J0220054 | JX657697 | JX560809 | TK147319 | TX | Dickens | 33°45′32″ N, 100°47′42″ W | 6/7/2008 | KC153486 |
| MPRV | AV J0220057 | JX657698 | JX560810 | TK147323 | TX | Dickens | 33°45′36″ N, 100°48′44″ W | 6/7/2008 | KC153487 |
| MPRV | AV J0210021 | JX657700 | JX560812 | TK147326 | TX | Dickens | 33°45′36″ N, 100°48′44″ W | 6/28/2008 | KC153484 |
| CTNV | AV A0400135 | DQ865244 | DQ865244 | TK84703 | TX | Dimmit | 28°20′58″ N, 99°25′39″ W | 7/19/1999 | FJ716220 |
| CTNV | AV C0410273 | JX657704 | JX289300 | TK100206 | TX | Dimmit | 28°20′23″ N, 99°25′22″ W | 3/13/2001 | ND |
| CTNV | AV C1170006 | JX657705 | JX289305 | TK100445 | TX | Dimmit | 28°18′37″ N, 99°20′49″ W | 10/4/2001 | ND |
| CTNV | AV A0400212 | DQ865245 | DQ865245 | TK84816 | TX | La Salle | 28°18′46″ N, 99°20′34″ W | 7/20/1999 | FJ716221 |
| CTNV | AV C0410246 | JX657708 | JX289299 | TK100126 | TX | La Salle | 28°21′12″ N, 99°27′59″ W | 3/11/2001 | ND |
| CTNV | AV D1030150 | JX657710 | JQ063088 | TK102318 | TX | La Salle | 28°21′12″ N, 99°27′59″ W | 10/5/2002 | ND |
WWAV, Whitewater Arroyo species complex virus; MPRV, Middle Pease River virus; CTNV, Catarina virus.
GenBank accession number.
The nucleotide sequences of AV H0380005, AV H0380014, AV H0380016, AV H0380020, AV 98490013, and the 6 CTNV strains were determined from RNA isolated from infected Vero E6 cells; the nucleotide sequences of the 12 other strains were determined from RNA isolated from kidney. The nucleotide sequence of the N protein gene of AV D1030150 and the nucleotide sequences of the GPC genes and N protein genes of AV H0380005, AV 98490013, AV A0400135, and AV A0400212 were determined in previous studies (Cajimat et al. 2007b, 2011; Milazzo et al. 2013); the other GPC and N protein gene sequences were determined in this study.
Museum of Texas Tech University.
NM, New Mexico; OK, Oklahoma; TX, Texas.
Otero County, Fort Bliss Military Base; Cimarron County, Black Mesa State Park and Nature Preserve; Motley County, a 160-ha ranch located 1.6 km south of the town of Flomot; Dickens County, a 350-ha ranch located 1.3 km north of the town of Afton; Dimmit and La Salle counties, 6151 ha on the Chaparral Wildlife Management Area.
Cytb, cytochrome b; ND, no data. The nucleotide sequences of the Cytb genes of 2 other southern plains woodrats (N. micropus) in this study were determined: TK83527, Otero County (32°28′16″ N, 105°42′05″ W), GenBank accession no. KC153475; TK69721, Ward County (31°38′51″ N, 102°49′23″ W), KC153485. Both of these animals were positive for antibody to WWAV strain AV9310135, negative for arenavirus, and negative for arenaviral RNA.
2.1. Antibody assay
Blood samples from the rodents from FBMB, FLOM, AFTN, and MSSP were tested in previous studies (Milazzo et al. 2010, Mauldin et al. 2013) for immunoglobulin G (IgG) to WWAV strain AV 9310135 (Fulhorst et al. 1996), using an ELISA (Bennett et al. 2000). The test antigen was prepared from Vero E6 cells infected with AV 9310135, the control (comparison) antigen was prepared from uninfected Vero E6 cells, and the working dilutions of the antigens were determined by box-titration against immune sera from white-throated woodrats (N. albigula) inoculated with AV 9310135 (Fulhorst et al. 2001). Serial fourfold dilutions (from 1:80 through 1:5,120) of each blood sample were tested against both antigens; IgG bound to antigen was detected by using a mixture of a goat anti-rat IgG peroxidase conjugate and goat anti-Peromyscus leucopus IgG peroxidase conjugate (Kirkegaard and Perry Laboratories, Gaithersburg, MD) in conjunction with the ABTS Microwell Peroxidase Substrate System (Kirkegaard and Perry Laboratories); optical densities (OD) were measured at 410 nm (reference, 490 nm); the adjusted OD (AOD) of a blood-antigen reaction was the OD of the well coated with the test antigen less the OD of the well coated with the control antigen; a sample was considered positive if the AOD at 1:80 was ≥ 0.250, the AOD at 1:320 was ≥ 0.250, and the sum of the AOD for the series of fourfold dilutions (from 1:80 through 1:5,120) was ≥ 0.750; and the titer of a positive sample was the highest dilution for which the AOD was ≥ 0.250.
2.2. Virus assay
Samples of kidney and spleen from the 477 rodents captured at FBMB, FLOM, AFTN, and MSSP were tested for arenavirus by cultivation in monolayers of Vero E6 cells (Fulhorst et al. 1996). Arenaviral antigen in infected Vero E6 cells was revealed by using an indirect fluorescent antibody test in which the primary antibody was a hyperimmune mouse ascitic fluid raised against WWAV strain AV 9310135.
2.3. Genetic characterization of arenaviruses isolated from woodrats
The nucleotide sequences of a 784- to 891-nt fragment of the 5′ half and 902- to 1157-nt fragment of the 3′ half of the small (S) genomic segments of arenaviruses AV H0380014, AV H0380016, and AV H0380020; the nucleotide sequence of a 1588-nt fragment of the 5′ half of the S genomic segment of CTNV strain AV D1030150; and the nucleotide sequences of a 586-nt fragment of the 5′ half and 587-nt fragment of the 3′ half of the S genomic segments of CTNV strains AV C0410273, AV C1170006, and AV C0410246 were determined from RNA isolated from monolayers of infected Vero E6 cells. The sequences from the 5′ half of the S segment included the entire region of the glycoprotein precursor (GPC) gene that encodes the G1; the sequences from the 3′ half of the S segment were from the nucleocapsid (N) protein gene; and the passage histories of the viruses in the inocula were Vero E6+1 or Vero E6+2. Reverse transcription of S segment RNA was done by using SuperScript III® Reverse Transcriptase (Invitrogen Corp., Carlsbad, CA) in conjunction with oligonucleotide 19C-cons (Cajimat et al. 2007a); the PCR assays used MasterTaq® Kit (5 PRIME, Inc., Gaithersburg, MD); and amplicons of the expected size were purified from agarose gel slices and sequenced directly.
2.4. Assays for arenaviral RNA
Samples of kidney from the antibody-positive, culture-negative rodents were tested for GPC gene RNA and N protein gene RNA. Briefly, total RNA was isolated from 30-45 mg of tissue; first-strand cDNA was synthesized by using SuperScript III® Reverse Transcriptase (Invitrogen Corp.) in conjunction with oligonucleotide 19C-cons (Cajimat et al. 2007a); the first-and second-round PCR assays used MasterTaq® Kit (5 PRIME, Inc.); the controls included kidney from a CTNV-infected, culture-positive southern plains woodrat and samples of kidney from antibody-negative, culture-negative southern plains woodrats; and amplicons of the expected size were purified from agarose gel slices and sequenced directly.
2.5. Genetic characterization of arenaviruses AV I0130006 and AV M0040017
The nucleotide sequences of a 3299-nt fragment of the S genomic segments of AV I0130006 (FLOM) and AV M0040017 (AFTN) were determined from RNA isolated from kidney, using first-strand cDNA generated with 19C-cons and methods published previously (Inizan et al. 2010). Each sequence extended from within the non-coding region (NCR) at the 5′ end of the S segment, through the GPC gene, intergenic region, and N protein gene, and into the NCR at the 3′ end of the S segment.
2.6. Genetic characterization of woodrats
The species identities of 21 arenavirus-infected woodrats in this study were confirmed by analyses of complete cytochrome-b (Cytb) gene sequences. The complete nucleotide sequences of the Cytb genes of woodrats TK77260, TK77270, TK84703, TK84816, and TK28731 were published previously (Cajimat et al. 2011, Edwards and Bradley 2002). The complete nucleotide sequences of the Cytb genes of the 16 other woodrats were determined in this study, using DNA isolated from liver, GoTaq® DNA Polymerase (Promega Corp., Madison, WI), oligonucleotides LGL 765 forward (Bickham et al. 1995) and LGL 766 reverse (Bickham et al. 2004), and methods published previously (Edwards and Bradley 2002).
2.7. Sequencing reactions and analysis
The nucleotide sequences of the purified amplicons were determined directly, using the BigDye® Terminator v.3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA). The products of the sequencing reactions were analyzed on an ABI PRISM® 3100-Avant™ or 3130-Avant™ Genetic Analyzer (Applied Biosystems).
2.8. Data analysis
The analyses of GPC gene sequences and N protein gene sequences included BCNV strains AV A0060209 (GenBank accession no. AF512833), AV 98470029 (AY924392), AV A0070039 (AY924391), and AV B0300052 (FJ907243 and FJ907244); BBTV, AV D0390174 (EF619035) and AV D0390324 (EF619036); CTNV, AV A0400135 (DQ865244) and AV A0400212 (DQ865245); RCTV, AV H0030026 (GQ903697); SKTV, AV D1000090 (EU123328); TAMV, W·10777 (AF512828) and AV 97140103 (EU486821); TTCV, AV D0150144 (EF619033) and AV D0390060 (EF619034); WWAV, AV 9310135 (AF228063); arenaviruses AV 96010024 (EU123331), AV 96010025 (EU486820), AV 96010151 (EU123330), AV 98490013 (FJ032026 and FJ032027), TVP·6038 (FJ719106 and FJ719107), and AV D1240007 (EU123329); Tacaribe virus, TRVL 11573 (M20304); and 14 South American Tacaribe serocomplex viruses – Allpahuayo virus, CLHP-2472 (AY012687); Amaparí virus, BeAn 70563 (AF512834); Chaparé virus, 200001071 (EU260463); Cupixi virus, BeAn 119303 (AF512832); Flexal virus, BeAn 293022 (AF512831); Guanarito virus, INH-95551 (AY129247); Junín virus, XJ13 (AY358023): Latino virus, MARU 10924 (AF512830); Machupo virus, Carvallo (AY129248); Oliveros virus, 3229-1 (U34248); Paraná virus, 12056 (AF485261); Pichindé virus, Co An 3739 (K02734); Pirital virus, VAV-488 (AF485262); and Sabiá virus, SPH 114202 (U41071). The alignments of GPC sequences and N protein sequences were constructed using the computer program CLUSTAL W1.7 (Thompson et al. 1994); the alignments of complete GPC gene sequences and complete N protein gene sequences were constructed manually, guided by the computer-generated amino acid sequence alignments; and sequence nonidentities were equivalent to uncorrected distances.
The analyses of Cytb gene sequences included southern plains woodrats (N. micropus) TK16501 (GenBank accession no. AF186824), TK28743 (KC812730), TK31643 (AF186822), TK49607 (AF376469), TK51947 (AF298844), TK51949 (AF298845), TK54820 (AF186825), TK84556 (AF186826), TK84557 (AF186827), and TK84761 (KC153488); white-throated woodrat (N. albigula) TK74854 (AF186803); eastern woodrat (N. floridana) TK52109 (AF186819); white-toothed woodrat (N. leucodon) TK49716 (AF186806); and Mexican woodrat (N. mexicana) TK90038 (AF294346). The Cytb gene sequences were aligned manually.
The Bayesian analyses were done with MRBAYES v3.1.2 (Huelsenbeck and Ronquist 2001) and programs in the computer software package PAUP* (Swofford 2003), using a GTR+I+G model of evolution with a site-specific gamma distribution and the following options in MRBAYES v3.1.2: two simultaneous runs of 4 Markov chains, five million generations (analyses of GPC and N protein gene sequences) or 10 million generations (analyses of Cytb gene sequences), and sample frequency = every 1,000th generation. The GTR+I+G model of evolution was selected based on the results of analyses done with MrModeltest (Nylander 2004); Pirital virus strain VAV-488 and Guanarito virus strain INH-95551 were the designated outgroups in the analyses of the GPC and N protein gene sequences, respectively; white-throated woodrat (N. albigula) TK74854 was the designated outgroup in the analyses of Cytb gene sequences; the first 1,000 trees from each analysis were discarded after review of likelihood scores, convergence statistics, and potential scale reduction factors; a majority-rules consensus tree was constructed from the remaining trees; and clade probability values were calculated a posteriori.
Differences among the amino acid sequences of the G1 of AV 98490013, AV H0380005, AV I0130006, AV M0040017, and CTNV strain AV A0400135 were scored favored, neutral, or disfavored, using substitution preferences for extracellular proteins (Betts and Russell 2003). Gaps in the alignments and disfavored differences were considered non-conservative.
Pairwise comparisons of GPC gene sequences by locality were restricted to the 576-nt region that encodes the G1. Similarly, pairwise comparisons of N protein gene sequences by locality were restricted to a 587-nt fragment near the 3′ end of the intergenic region of the S genomic segment.
3. Results
Arenavirus was isolated from 4 (66.7%) of 6 antibody-positive woodrats from FBMB (Table 1), none of 81 other rodents from FBMB, and none of 390 rodents from FLOM, AFTN, or MSSP. Arenaviral RNA was found in 12 (66.7%) of 18 antibody-positive, culture-negative rodents: 1 (50.0%) of 2 woodrats from FBMB, 4 (80.0%) of 5 woodrats from FLOM, 7 of 7 woodrats and none of 2 cotton rats (Sigmodon hispidus) from AFTN, none of 2 woodrats from MSSP. The results of the Bayesian analyses of the nucleotide sequences of a 587-nt fragment of the N protein genes of 23 arenaviruses associated with N. micropus (Table 2) and 34 other Tacaribe serocomplex viruses (see “Data analysis”) indicated that the 5 arenaviruses associated with woodrats captured on FBMB are monophyletic, the 11 arenaviruses associated with woodrats captured at FLOM and AFTN are monophyletic, and the arenaviruses associated with woodrats captured on FBMB are phylogenetically distinct from the arenaviruses associated with woodrats captured at FLOM and AFTN. The results of the Bayesian analyses of Cytb gene sequence data indicated that woodrat TK28731 from BMSP; the antibody-positive woodrats from FBMB, FLOM, AFTN, and MSSP; and the woodrats from the CWMA were N. micropus.
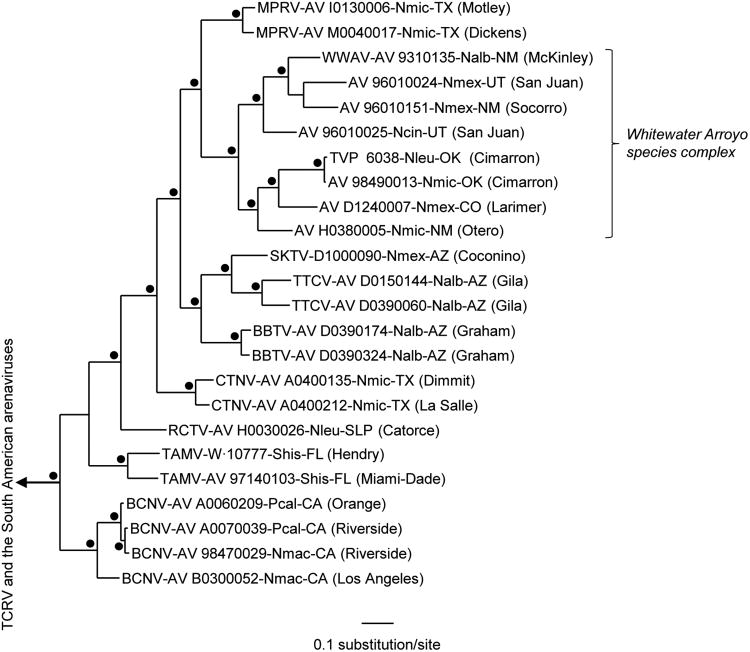
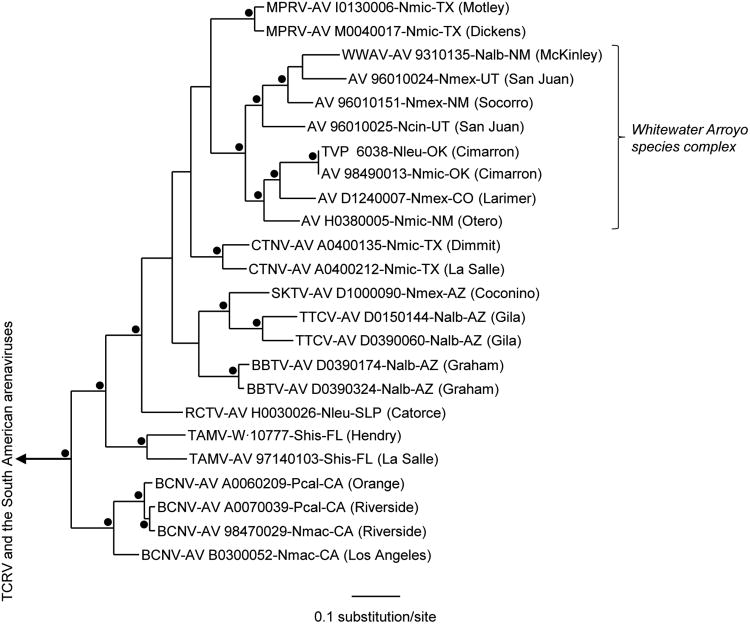
Nonidentities among the complete nucleotide sequences of the GPC genes and among the complete nucleotide sequences of the N protein genes of AV 98490013 (BMSP), AV H0380005 (FBMB), AV I0130006 (FLOM), AV M0040017 (AFTN), and CTNV strain AV A0400135 (CWMA) ranged from 7.0% (AV I0130006 and AV M0040017) to 33.3% (AV I0130006 and AV 98490013) and from 3.2% (AV I0130006 and AV M0040017) to 23.1% (AV I0130006 and AV H0380005), respectively. The Bayesian analyses of complete GPC gene sequences (Figure 2) indicated that AV I0130006 and AV M0040017 are monophyletic and phylogenetically more closely related to the 8 Whitewater species complex viruses (Cajimat et al. 2008) than to other Tacaribe serocomplex viruses. The results of the Bayesian analyses of complete N protein gene sequences (Figure 3) also indicated that AV I0130006 and AV M0040017 are monophyletic but did not solve the relationship between AV I0130006 (or AV M0040017) and the Tacaribe serocomplex viruses found in association with woodrats (Neotoma spp.) captured in Arizona, Colorado, New Mexico, Oklahoma, Utah, southern Texas, or San Luis Potosí. We note that many of the topographical differences between the GPC gene tree (Figure 2) and N protein gene tree (Figure 3) were not supported by clade support values calculated a posteriori; notably, many of the North American viruses in the N protein gene tree could collapse into unresolved polytomies.
Figure 2.
Phylogenetic relationships among the North American Tacaribe serocomplex viruses based on Bayesian analyses of complete glycoprotein precursor gene sequences. The length of the scale bar is equivalent to 0.1 substitution per site. A black dot at a node indicates that the probability values in support of the clade were ≥ 0.95. The branch labels include (in the following order) virus species, strain, host species, state (and county or municipality). BBTV, Big Brushy Tank virus; BCNV, Bear Canyon virus; CTNV, Catarina virus; MPRV, Middle Pease River virus; RCTV, Real de Catorce virus; SKTV, Skinner Tank virus; TAMV, Tamiami virus; TCRV, Tacaribe virus; TTCV, Tonto Creek virus; WWAV, Whitewater Arroyo virus. Nalb, Neotoma albigula (white-throated woodrat); Ncin, N. cinerea (bushy-tailed woodrat); Nleu, N. leucodon (white-toothed woodrat); Nmac, N. macrotis (large-eared woodrat); Nmex, N. mexicana (Mexican woodrat); Nmic, N. micropus (southern plains woodrat); Peromyscus californicus (California mouse); Shis, S. hispidus (hispid cotton rat). AZ, Arizona; CA, California; CO; Colorado; FL, Florida; NM, New Mexico; OK, Oklahoma; SLP, San Luis Potosí; TX, Texas; UT, Utah.
Figure 3.
Phylogenetic relationships among the North American Tacaribe serocomplex viruses based on Bayesian analyses of complete nucleocapsid protein gene sequences. The length of the scale bar is equivalent to 0.1 substitution per site. Nodal support and branch labels are as in Figure 2.
The lengths of the GPC of AV I0130006 and AV M0040017 were comparable to the lengths of the GPC of the 22 other North American Tacaribe serocomplex viruses (Table 3). The amino acid sequence of the GPC of AV I0130006 was 6.0% different from the amino acid sequence of the GPC of AV M0040017; nonidentities between the amino acid sequences of the GPC of AV I0130006 and AV M0040017 and the amino acid sequences of the GPC of the 8 Whitewater Arroyo species complex viruses ranged from 30.6% (AV M0040017 and AV 96010151) to 34.6% (AV I0130006 and AV 98490013); and nonidentities between the amino acid sequences of the GPC of AV I0130006 and AV M0040017 and the amino acid sequences of the GPC of the other North American Tacaribe serocomplex viruses ranged from 27.9% to 37.8% (Table 4).
Table 3.
North American arenaviruses included in the analyses of nucleotide and amino acid sequence data.
| Length of peptide or protein (aa)b | |||||||
|---|---|---|---|---|---|---|---|
|
|
|||||||
| Speciesa | Strain | Genbank accession no(s). | |||||
| GPC | SP | G1 | G2 | N protein | |||
| MPRV | AV I0130006 | JX560798 | 484 | 58 | 192 | 234 | 562 |
| MPRV | AV M0040017 | JX560799 | 484 | 58 | 192 | 234 | 562 |
| BCNV | AV A0060209 | AF512833 | 483 | 58 | 191 | 234 | 562 |
| BCNV | AV 98470029 | AY924392 | 483 | 58 | 191 | 234 | 562 |
| BCNV | AV A0070039 | AY924391 | 483 | 58 | 191 | 234 | 562 |
| BCNV | AV B0300052 | FJ907243, FJ907244 | 483 | 58 | 191 | 234 | 562 |
| BBTV | AV D0390174 | EF619035 | 485 | 58 | 193 | 234 | 562 |
| BBTV | AV D0390324 | EF619036 | 485 | 58 | 193 | 234 | 562 |
| CTNV | AV A0400135 | DQ865244 | 484 | 58 | 192 | 234 | 562 |
| CTNV | AV A0400212 | DQ865245 | 484 | 58 | 192 | 234 | 562 |
| RCTV | AV H0030026 | GQ903697 | 483 | 58 | 191 | 234 | 562 |
| SKTV | AV D1000090 | EU123328 | 481 | 58 | 189 | 234 | 562 |
| TAMV | AV 97140103 | EU486821 | 485 | 58 | 193 | 234 | 562 |
| TAMV | W·10777 | AF512828 | 485 | 58 | 193 | 234 | 562 |
| TTCV | AV D0150144 | EF619033 | 484 | 58 | 192 | 234 | 562 |
| TTCV | AV D0390060 | EF619034 | 485 | 58 | 193 | 234 | 562 |
| WWAV | AV 9310135 | AF228063 | 480 | 58 | 188 | 234 | 562 |
| WWASC | AV 96010024 | EU123331 | 480 | 58 | 188 | 234 | 562 |
| WWASC | AV 96010025 | EU486820 | 480 | 58 | 188 | 234 | 562 |
| WWASC | AV 96010151 | EU123330 | 483 | 58 | 191 | 234 | 562 |
| WWASC | AV 98490013 | FJ032026, FJ032027 | 482 | 58 | 190 | 234 | 562 |
| WWASC | TVP·6038 | FJ719106, FJ719107 | 482 | 58 | 190 | 234 | 562 |
| WWASC | AV D1240007 | EU123329 | 484 | 58 | 192 | 234 | 562 |
| WWASC | AV H0380005 | EU910959 | 484 | 58 | 192 | 234 | 562 |
MPRV, Middle Pease River virus; BCNV, Bear Canyon virus; BBTV, Big Brushy Tank virus; CTNV, Catarina virus; RCTV, Real de Catorce virus; SKTV, Skinner Tank virus; TAMV, Tamiami virus; TTCV, Tonto Creek virus; WWAV, Whitewater Arroyo virus. MPRV, BBTV, CTNV, RCTV, SKTV, and TTCV are provisional species in the family Arenaviridae, genus Arenavirus. Arenaviruses AV 96010024, AV 96010025, AV 96010151, AV 98490013, TVP·6038, AV D1240007, and AV H0380005 are members of the Whitewater Arroyo species complex (WWASC) (Cajimat et al. 2008).
GPC, glycoprotein precursor; SP, signal peptide; N protein, nucleocapsid protein.
Table 4.
Nonidentities among the amino acid sequences of the complete glycoprotein precursors and among the amino acid sequences of the complete nucleocapsid proteins of 24 North American Tacaribe serocomplex viruses.a
| Glycoprotein precursor (% sequence nonidentity) | |||||
|---|---|---|---|---|---|
|
|
|||||
| Virus(es)b | AV I0130006 | AV M0040017 | CTNVb | WWAVc | Otherd |
| AV I0130006 | -- | 6.0 | 30.6–31.3 | 30.8–34.6 | 27.9–37.6 |
| AV M0040017 | 0.5 | -- | 31.1–31.5 | 30.6–33.6 | 28.1–37.8 |
| CTNVb | 11.7–12.1 | 11.4–11.7 | -- | 31.6–35.4 | 26.7–36.8 |
| WWAVc | 12.5–14.8 | 12.3–14.9 | 11.4–14.6 | -- | 29.6–41.6 |
| Otherd | 11.9–19.9 | 11.6–19.8 | 9.8–18.5 | 10.9–21.0 | -- |
| Nucleocapsid protein (% sequence nonidentity) | |||||
Nonidentities (uncorrected p-model distances) among the glycoprotein precursors (GPC) and among the nucleocapsid (N) proteins are listed above and below the diagonal, respectively.
CTNV, Catarina virus strains AV A0400135 and AV A0400212.
WWAV, Whitewater Arroyo virus prototype strain AV 9310135 and Whitewater Arroyo species complex viruses AV H0380005, AV 98490013, TVP·6038, AV 96010024, AV 96010025, AV 96010151, and AV D1240007 (Cajimat et al. 2008). Nonidentities among the amino sequences of the GPC and among the amino acid sequences of the N proteins of the 8 Whitewater Arroyo species complex viruses ranged from 1.0% (TVP·6038 and AV 98490013) to 25.8% (AV 96010024 and AV D1240007) and 0.2% (TVP·6038 and AV 98490013) to 10.5% (AV 96010024 and AV 96010025), respectively.
The other North American viruses were Big Brushy Tank virus (BBTV) strains AV D0390174 and AV D0390324; Bear Canyon virus (BCNV) strains AV A0060209, AV A0070039, AV 98470029, and AV B0300052; RCTV, Real de Catorce virus (RCTV) strain AV H0030026; Skinner Tank virus (SKTV) strain AV D1000090; Tamiami virus (TAMV) strains W·10777 and AV 97140103; and Tonto Creek virus (TTCV) strains AV D0150144 and AV D0390060. Nonidentities among the amino acid sequences of the GPC and among the amino acid sequences of the N proteins of strains of different species in the “Other” category ranged from 19.5% (SKTV strain AV D1000090 and TTCV strain AV D0390060) to 37.3% (BBTV strain AV D0390324 and TAMV strain AV 97140103) and 9.4% (SKTV strain AV D1000090 and TTCV strains AV D0150144 and AV D0390060) to 21.7% (BCNV strain AV A0060209 and TAMV strain W-10777), respectively.
The lengths of the N proteins of AV I0130006 and AV M0040017 were identical to the lengths of the N proteins of the 22 other North American Tacaribe serocomplex viruses (Table 3). The amino acid sequence of the N protein of AV I0130006 was 0.5% different from the amino acid sequence of the N protein of AV M0040017; nonidentities between the amino acid sequences of the N proteins of AV I0130006 and AV M0040017 and the amino acid sequences of the N proteins of the 8 Whitewater Arroyo species complex viruses ranged from 12.3% (AV M0040017 and TVP·6038) to 14.9% (AV M0040017 and AV 96010024); and nonidentities between the amino acid sequences of the N proteins of AV I0130006 and AV M0040017 and the amino acid sequences of the N proteins of the other North American Tacaribe serocomplex viruses ranged from 11.6% to 19.9% (Table 4).
The GPC of AV I0130006, AV M0040017, and the other North American Tacaribe serocomplex viruses contained a potential signal peptidase cleavage site after residue 58 (SCS58↓); the GPC of AV I0130006 and AV M0040017 contained a potential SKI-1/S1P cleavage site after residue 250 (RKLQ250↓); and the GPC of the other North American Tacaribe serocomplex viruses contained a potential SKI-1/S1P cleavage site within the region flanked by residues 246 and 252 (Cajimat et al. 2011). Accordingly, the predicted lengths of the G1 of the North American arenaviruses ranged from 188 to 193 aa (Table 3).
Nonidentities among the amino acid sequences of the G1 of AV 98490013, AV H0380005, AV I0130006, AV M0040017, and CTNV strain AV A0400135 ranged from 10.9% (AV I0130006 and AV M0040017) to 55.6% (AV 98490013 and AV A0400135). The prevalence of non-conservative differences among the amino acid sequences of the G1 of these 5 viruses ranged from 1.0% (AV I0130006 and AV M0040017) to 11.5% (AV 98490013 and AV A0400135), with a median of 7.3%; and the non-conservative differences were distributed along the primary structure of the G1, from residue 18 through residue 188.
By locality, nonidentities among the nucleotide sequences of the GPC genes and among the nucleotide sequences of the N protein genes of the viruses from FBMB, FLOM, AFTN, and the CWMA were as high as 15.1% (FBMB) and 10.2% (CWMA), respectively (Table 5). Similarly, nonidentities among the amino acid sequences of the G1 and among the amino acid sequences of the N proteins of viruses from the same locality were as high as 15.6% (FBMB) and 5.1% (FBMB), respectively (Table 5).
Table 5.
Nonidentities among the nucleotide sequences and among the predicted amino acid sequences of 22 arenaviruses naturally associated with Neotoma micropus, by locality.
| Sequence nonidentitiesa | ||||||
|---|---|---|---|---|---|---|
|
|
||||||
| Virusb | Localityc | Number of strains | GPC, ntd | G1, aae | N protein, ntf | N protein, aag |
| WWAV | FBMB | 5 | 0.0--15.1 | 0.0--15.6 | 0.0--8.5 | 0.0--5.1 |
| MPRV | FLOM | 4 | 0.2--1.7 | 0.0--1.0 | 0.2--1.0 | 0.0--0.5 |
| MPRV | AFTN | 7 | 0.0--1.0 | 0.0--1.6 | 0.0--1.9 | 0.0--1.5 |
| CTNV | CWMA | 6 | 0.0--12.0 | 0.0--10.9 | 0.2--10.2 | 0.0--3.6 |
Percent (%) nonidentity.
WWAV, Whitewater Arroyo species complex viruses associated with N. micropus on Fort Bliss Military Base; MPRV, Middle Pease River virus; CTNV, Catarina virus.
FBMB, Fort Bliss Military Base in Otero County, New Mexico; FLOM, a ranch near the town of Flomot in Motley County, Texas; AFTN, a ranch near the town of Afton in Dickens County, Texas; CWMA, Chaparral Wildlife Management Area in Dimmit and La Salle counties, Texas.
Nucleotide sequences of the 576-nt fragment of the glycoprotein precursor (GPC) gene that encodes the G1 (glycoprotein). Nonidentities between the GPC gene sequences of the MPRV strains from FLOM and MPRV strains from AFTN ranged from 0/576 to 66/576 (11.5%).
Nonidentities between the amino acid sequences of the G1 of the MPRV strains from FLOM and MPRV strains from AFTN ranged from 0/192 to 25/192 (13.0%).
Nucleotide sequences of a 587-nt fragment of the N protein gene, 593-nt downstream from the intergenic region of the small genomic segment. Nonidentities between the N protein gene sequences of the MPRV strains from FLOM and MPRV strains from AFTN ranged from 0/587 to 23/587 (3.9%).
Nonidentities between the sequences of a 195-aa fragment of the N proteins of the MPRV strains from FLOM and MPRV strains from AFTN ranged from 0/595 to 3/195 (1.5%).
4. Discussion
The pairwise comparisons of complete GPC gene sequences and complete N protein gene sequences in this study revealed a high level of genetic diversity among Tacaribe serocomplex viruses associated with N. micropus in the United States, comparable to the level of genetic diversity among Tacaribe serocomplex viruses associated with N. albigula in Arizona and New Mexico (Milazzo et al. 2008) as well as the level of genetic diversity among Tacaribe serocomplex viruses associated with N. mexicana in Arizona, Colorado, New Mexico, and Utah (Cajimat et al. 2008). The results of Bayesian analyses of complete GPC gene sequences and complete N protein gene sequences in a previous study (Cajimat et al. 2011) and this study indicated that AV 98490013 (BMSP) and AV H0380005 (FBMB) should be included in the Whitewater Arroyo species complex. The results of the Bayesian analyses of GPC and N protein gene sequences in this study also indicated that the arenaviruses associated with N. micropus at FLOM and AFTN are monophyletic and phylogenetically closely related to the Whitewater Arroyo species complex viruses.
In a previous study (Milazzo et al. 2008), nonidentities among the amino acid sequences of the GPC and among the amino acid sequences of the N proteins of strains of different South American arenaviral species were as low as 15.8% (Allpahuayo virus strain CLHP-2472 and Flexal virus strain BeAn 293022) and 11.9% (Junín virus strain XJ13 and Machupo virus strain 9530537), respectively. Collectively, the results of the Bayesian analyses of complete GPC gene sequences, pairwise comparisons of complete GPC sequences, and pairwise comparisons of complete N protein sequences in this study indicate that AV I0130006 and AV M0040017 are strains of a novel arenaviral species, tentatively named Middle Pease River virus (MPRV). We note that the MPRV-infected woodrats from FLOM were captured in close proximity to the Middle Pease River.
The most recent report of the International Committee on Taxonomy of Viruses (Salvato et al. 2012) indicated that strains of different species in the Arenaviridae should exhibit at least a 12.0% difference in pairwise comparisons of complete N protein sequences, which is slightly greater than the level of nonidentity between the sequences of the N proteins of MPRV strains AV I0130006 and AV M0040017 and the sequences of the N proteins of BBTV strain AV D0390174 and CTNV strain AV A0400212 (Table 4). However, the numerical basis for the 12.0% cut-off is not known with certainty to the authors of this study; none of the criteria for species demarcation within the Arenaviridae presently include consideration of non-identities between GPC sequences or phylogenetic relationships among members of the Arenaviridae; and the most recent report of the International Committee on Taxonomy of Viruses did not acknowledge CTNV or any of 4 other North American Tacaribe serocomplex viruses described in the peer-reviewed scientific literature in 2007-2012 (Cajimat et al. 2007b, 2008, 2012; Inizan et al. 2010; Milazzo et al. 2008). The authors of this study suggest that a revision of the criteria for species demarcation within the Arenaviridae is required. This revision should consider differences in ecology, cross-neutralization tests, N protein sequences, GPC sequences, and other phenetic characteristics in the context of phylogenetic relationships estimated from complete GPC gene sequences and complete N protein gene sequences.
Together, the results of the assays for arenavirus and assays for anti-arenavirus IgG in this study suggest that the southern plains woodrat is the principal host of MPRV at AFTN and FLOM, and the principal host of the Whitewater Arroyo species complex virus found on FBMB. The results of the serological assays also suggest that the hispid cotton rat (S. hispidus) at AFTN is a natural host of MPRV or another Tacaribe serocomplex virus that is cross-reactive with WWAV strain AV 9310135 in ELISA. Alternatively, the serological results for the “antibody-positive” cotton rats were falsely positive. We note that the antibody titers in the “antibody-positive” cotton rats were only 320 whereas the antibody titers in 10 (83.3%) of the 12 antibody-positive woodrats from AFTN and FLOM were ≥5120; the assays for arenavirus in the “antibody-positive” cotton rats and 68 other cotton rats captured at AFTN were negative; and the assays for N protein gene RNA in the “antibody-positive” cotton rats were negative.
The principal host relationships of some South American Tacaribe serocomplex viruses appear to represent a long-term shared evolutionary relationship between the Arenaviridae and the Cricetidae, subfamily Sigmodontinae (Bowen et al. 1997). Evidence for this long-standing relationship includes the present-day association of phylogenetically closely related arenaviral species with phylogenetically closely related members of the Sigmodontinae, for example – Junín virus with Calomys musculinus in Argentina (Mills et al. 1992) and Machupo virus with a Calomys species in Bolivia (Johnson et al. 1966, Salazar-Bravo et al. 2002). Hypothetically, the principal host relationships of some North American Tacaribe serocomplex viruses represent a long-term shared evolutionary relationship between the Arenaviridae and the Cricetidae, subfamily Neotominae, genus Neotoma.
Specific knowledge of the natural host relationships of many of the North American Tacaribe serocomplex viruses is limited to the results of assays for arenavirus or arenaviral RNA in tissues from a small number of wild-caught rodents (Cajimat et al. 2011). As such, some of the virus-rodent pairings in Figure 2 and Figure 3 may be a consequence of contemporary, interspecific virus transmission rather than representative of an ancient relationship between the Arenaviridae and specific phylogenetic lineages (species) within the genus Neotoma. Clearly, the ecologies (principal host relationships) of the Whitewater Arroyo species complex viruses and many of the other North American Tacaribe serocomplex viruses need to be elucidated before serious consideration of the origins of the present-day associations between N. micropus and MPRV, N. micropus and CTNV, and N. micropus and members of the Whitewater Arroyo species complex.
The dominant neutralizing epitopes on an arenavirion are associated with G1 (Buchmeier et al. 1981), and antibody-mediated neutralization in vitro can vary from strain to strain within an arenavirus species (Jahrling and Peters 1984, Parekh and Buchmeier 1986). The high prevalence of non-conservative differences among the amino acid sequences of the G1 of AV 98490013, AV H0380005, MPRV strain AV I0130006 (or AV M0040017), and CTNV strain AV A0400135 suggests that there may be significant differences among these viruses in cross-neutralization tests done in vitro.
Previous studies revealed a high level of diversity among the nucleotide sequences of the N protein genes of CTNV strains associated with N. micropus on the CWMA (Fulhorst et al. 2002, Milazzo et al. 2013) and a high level of genetic diversity in N. micropus on the CWMA (Méndez-Harclerode et al. 2007). This study revealed a similar level of diversity among the nucleotide sequences of the N protein genes of arenaviruses associated with N. micropus on FBMB, and even higher levels of diversity among the nucleotide sequences of the GPC genes of arenaviruses associated with N. micropus on the CWMA and among the nucleotide sequences of the GPC genes of arenaviruses associated with N. micropus on FBMB (Table 5). Hypothetically, the co-occurrence of multiple genetic forms of CTNV on the CWMA is a consequence of commingling of allopatric N. micropus populations, each infected with a unique genetic form of CTNV (Fulhorst et al. 2002). Similarly, the co-occurrence of multiple genetic forms of the arenavirus associated with N. micropus on FBMB may be a consequence of mixing of allopatric N. micropus populations.
Acknowledgments
The first 3 authors (MNBC, MLM, and MRM) contributed equally to this study. John D. Hanson, Sheri Westerman-Ayers, Ryan Duplechin, Megan Corley-Keith, and Cody W. Thompson (Texas Tech University) captured the rodents from Dickens County. Robert J. Baker and Heath Garner facilitated the loan of tissues from the Museum of Texas Tech University. This study was financially supported by grant AI-41435 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Maria N. B. Cajimat, Email: nbcajirm@utmb.edu.
Mary Louise Milazzo, Email: mamilazz@utmb.edu.
Matthew R. Mauldin, Email: matt.mauldin@ttu.edu.
Robert D. Bradley, Email: robert.bradley@ttu.edu.
References
- Bennett SG, Milazzo ML, Webb JP, Jr, Fulhorst CF. Arenavirus antibody in rodents indigenous to coastal southern California. Am J Trop Med Hyg. 2000;62:626–630. doi: 10.4269/ajtmh.2000.62.626. [DOI] [PubMed] [Google Scholar]
- Betts MJ, Russell RB. Amino acid properties and consequences of substitutions. In: Barnes MR, Gray IC, editors. Bioinformatics for Geneticists. Wiley; 2003. pp. 290–316. [Google Scholar]
- Bickham JW, Wood CC, Patton JC. Biogeographic implications of cytochrome b sequences and allozymes in sockeye (Oncorhynchus nerka) J Hered. 1995;86:140–144. doi: 10.1093/oxfordjournals.jhered.a111544. [DOI] [PubMed] [Google Scholar]
- Bickham JW, Patton JC, Schlitter DA, Rautenbach IL, Honeycutt RL. Molecular phylogenetics, karyotypic diversity, and partition of the genus Myotis (Chiroptera: Vespertilionidae) Mol Phylogenet Evol. 2004;33:333–338. doi: 10.1016/j.ympev.2004.06.012. [DOI] [PubMed] [Google Scholar]
- Bowen MD, Peters CJ, Nichol ST. Phylogenetic analysis of the Arenaviridae: patterns of virus evolution and evidence for cospeciation between arenaviruses and their rodent hosts. Mol Phylogenet Evol. 1997;8:301–316. doi: 10.1006/mpev.1997.0436. [DOI] [PubMed] [Google Scholar]
- Buchmeier MJ, Lewicki HA, Tomori O, Oldstone MBA. Monoclonal antibodies to lymphocytic choriomeningitis and Pichinde viruses: generation, characterization, and cross-reactivity with other arenaviruses. Virology. 1981;113:73–85. doi: 10.1016/0042-6822(81)90137-9. [DOI] [PubMed] [Google Scholar]
- Cajimat MNB, Milazzo ML, Hess BD, Rood MP, Fulhorst CF. Principal host relationships and evolutionary history of the North American arenaviruses. Virology. 2007a;367:235–243. doi: 10.1016/j.virol.2007.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajimat MNB, Milazzo ML, Bradley RD, Fulhorst CF. Catarina virus, an arenaviral species principally associated with Neotoma micropus (southern plains woodrat) in Texas. Am J Trop Med Hyg. 2007b;77:732–736. [PubMed] [Google Scholar]
- Cajimat MNB, Milazzo ML, Borchert JN, Abbott KD, Bradley RD, Fulhorst CF. Diversity among Tacaribe serocomplex viruses (family Arenaviridae) naturally associated with the Mexican woodrat (Neotoma mexicana) Virus Res. 2008;133:211–217. doi: 10.1016/j.virusres.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajimat MNB, Milazzo ML, Haynie ML, Hanson JD, Bradley RD, Fulhorst CF. Diversity and phylogenetic relationships among the North American Tacaribe serocomplex viruses (family Arenaviridae) Virology. 2011;421:87–95. doi: 10.1016/j.virol.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajimat MNB, Milazzo ML, Bradley RD, Fulhorst CF. Ocozocoautla de Espinosa virus and hemorrhagic fever, Mexico. Emerg Infect Dis. 2012;18:401–405. doi: 10.3201/eid1803.111602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calisher CH, Nabity S, Root JJ, Fulhorst CF, Beaty BJ. Transmission of an arenavirus in white-throated woodrats (Neotoma albigula) in southeastern Colorado, 1995-1999. Emerg Infect Dis. 2001;7:397–402. doi: 10.3201/eid0703.010305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards CW, Bradley RD. Molecular systematics of the genus Neotoma. Mol Phylogenet Evol. 2002;25:489–500. doi: 10.1016/s1055-7903(02)00294-4. [DOI] [PubMed] [Google Scholar]
- Fulhorst CF, Bowen MD, Ksiazek TG, Rollin PE, Nichol ST, Kosoy MY, Peters CJ. Isolation and characterization of Whitewater Arroyo virus, a novel North American arenavirus. Virology. 1996;224:114–120. doi: 10.1006/viro.1996.0512. [DOI] [PubMed] [Google Scholar]
- Fulhorst CF, Milazzo ML, Bradley RD, Peppers LL. Experimental infection of Neotoma albigula (Muridae) with Whitewater Arroyo virus (Arenaviridae) Am J Trop Med Hyg. 2001;65:147–151. doi: 10.4269/ajtmh.2001.65.147. [DOI] [PubMed] [Google Scholar]
- Fulhorst CF, Milazzo ML, Carroll DS, Charrel RN, Bradley RD. Natural host relationships and genetic diversity of Whitewater Arroyo virus in southern Texas. Am J Trop Med Hyg. 2002;67:114–118. doi: 10.4269/ajtmh.2002.67.114. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist FR. MRBAYES: Bayesian inference of phylogeny. Biometrics. 2001;17:754–756. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Inizan CC, Cajimat MNB, Milazzo ML, Barragán-Gomez A, Bradley RD, Fulhorst CF. Genetic evidence for a Tacaribe serocomplex virus (family Arenaviridae) in Mexico. Emerg Infect Dis. 2010;16:1007–1010. doi: 10.3201/eid1606.091648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahrling PB, Peters CJ. Passive antibody therapy of Lassa fever in cynomologus monkeys: importance of neutralizing antibody and Lassa virus strain. Infect Immun. 1984;44:528–533. doi: 10.1128/iai.44.2.528-533.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KM, Kuns ML, Mackenzie RB, Webb PA, Yunkers CE. Isolation of Machupo virus from wild rodent Calomys callosus. Am J Trop Med Hyg. 1966;15:103–106. doi: 10.4269/ajtmh.1966.15.103. [DOI] [PubMed] [Google Scholar]
- Mauldin MR, Corley MS, Milazzo ML, Hanson JD. Assessment of hantavirus and arenavirus prevalence in Dickens County. Texas Texas J Sci. 2013 in press. [Google Scholar]
- Méndez-Harclerode FM, Strauss RE, Fulhorst CF, Milazzo ML, Ruthven DC, III, Bradley RD. Molecular evidence for high levels of intrapopulation genetic diversity in woodrats (Neotoma micropus) J Mammal. 2007;88:360–370. doi: 10.1644/05-MAMM-A-377R1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milazzo ML, Cajimat MNB, Haynie ML, Abbott KD, Bradley RD, Fulhorst CF. Diversity among Tacaribe serocomplex viruses (family Arenaviridae) naturally associated with the white-throated woodrat (Neotoma albigula) in the southwestern United States. Vector Borne Zoonotic Dis. 2008;8:523–530. doi: 10.1089/vbz.2007.0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milazzo ML, Barragán-Gomez A, Hanson JD, Estrada-Franco JG, Arellano E, González-Cózatl FX, Fernández-Salas I, Ramirez-Aguilar F, Rogers DS, Bradley RD, Fulhorst CF. Antibodies to Tacaribe serocomplex viruses (family Arenaviridae, genus Arenavirus) in cricetid rodents from New Mexico, Texas, and Mexico. Vector-Borne Zoonotic Dis. 2010;10:629–637. doi: 10.1089/vbz.2009.0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milazzo ML, Campbell GL, Fulhorst CF. Novel arenavirus infection in humans, United States. Emerg Infect Dis. 2011;17:1417–1420. doi: 10.3201/eid1708.110285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milazzo ML, Amman BR, Cajimat MNB, Méndez-Harclerode FM, Suchecki JR, Hanson JD, Haynie ML, Baxter BD, Milazzo C, Jr, Carroll SA, Carroll DS, Ruthven DC, III, Bradley RD, Fulhorst CF. Ecology of Catarina virus (family Arenaviridae) in southern Texas, 2001-2004. Vector-Borne Zoonotic Dis. 2013;13:50–59. doi: 10.1089/vbz.2012.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills JN, Ellis BA, McKee KT, Jr, Calderon GE, Maiztegui JI, Nelson GO, Ksiazek TG, Peters CJ, Childs JE. A longitudinal study of Junin virus activity in the rodent reservoir of Argentine hemorrhagic fever. Am J Trop Med Hyg. 1992;47:749–763. doi: 10.4269/ajtmh.1992.47.749. [DOI] [PubMed] [Google Scholar]
- Musser GG, Carleton MD. Family Cricetidae. In: Wilson DE, Reeder DM, editors. Mammal Species of the World: A Taxonomic and Geographic Reference. 3rd. Johns Hopkins University Press; Baltimore: 2005. pp. 955–1189. [Google Scholar]
- Nylander JAA. MrModeltest v2 Program distributed by the author. Vol. 2 Evolutionary Biology Centre, Uppsala University; 2004. [Google Scholar]
- Parekh BS, Buchmeier MJ. Proteins of lymphocytic choriomeningitis virus: antigenic topography of the viral glycoproteins. Virology. 1986;153:168–178. doi: 10.1016/0042-6822(86)90020-6. [DOI] [PubMed] [Google Scholar]
- Patterson BD, Ceballos G, Sechrest W, Tognelli MF, Brooks T, Luna L, Ortega P, Salazar I, Young BE. Digital distribution maps of the mammals of the western hemisphere, version 3.0. NatureServe; Arlington, Virginia, USA: 2007. [Google Scholar]
- Salazar-Bravo J, Dragoo JW, Bowen MD, Peters CJ, Ksiazek TG, Yates TL. Natural nidality in Bolivian hemorrhagic fever and the systematics of the reservoir species. Infect Genet Evol. 2002;1:191–199. doi: 10.1016/s1567-1348(02)00026-6. [DOI] [PubMed] [Google Scholar]
- Salvato MS, Clegg JCS, Buchmeier MJ, Charrel RN, Gonzalez JP, Lukashevich IS, Peters CJ, Romanowski V. Family Arenaviridae. In: King AMQ, Lefkowitz EJ, Adams MJ, Carstens EB, editors. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses (ICTV) Academic Press; 2012. pp. 715–723. [Google Scholar]
- Swofford DL. PAUP* Phylogenetic Analysis Using Parsimony (*and Other Methods), version 4. Sunderland, Massachusetts: Sinauer Associates; 2003. [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W (1.7): improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choices. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]