Figure 1. BCL6 represses LITAF transcription in B cells.
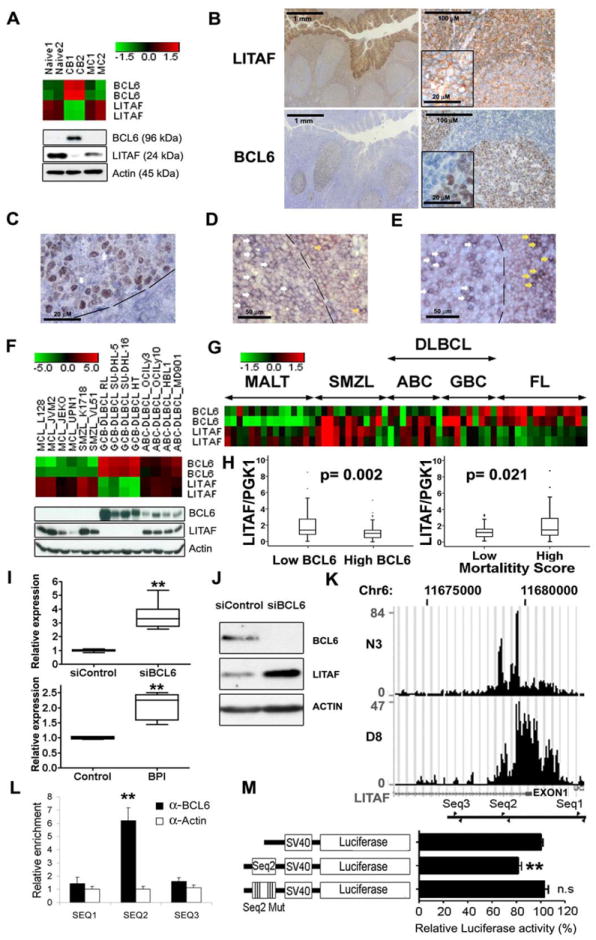
LITAF and BCL6 expression were assessed by analysis of gene expression microarrays (heatmap of RMA normalized log2 transformed values, two probe sets per gene) (A, upper panel) Western blotting (A, lower panel), immunohistochemistry (brown) (B) and double immunostaining (BCL6 brown and LITAF blue) (C) in reactive human tonsils. The white arrow in C points to a cell strongly expressing LITAF that is negative for BCL6 staining inside the GC, delimited by a discontinuous line. Details of higher magnification of the cells are embedded in two micrographs in B. Double staining for LITAF in red and CD20 (D) or CD3 (E) in blue showed that some B cells as well as T cells express LITAF inside (white arrows) and outside (yellow arrows) the GC. (F) Heatmap of microarray gene expression data (upper panel) and protein expression assessed by Western blot (lower panel) of LITAF and BLC6 in cell lines of mantle cell lymphoma (MCL), splenic marginal zone lymphoma (SMZL), diffuse large B-cell lymphoma of the GC subtype (GCB-DLBCL) and of the activated B-cell-like subtype (ABC-DLBCL). LITAF and BCL6 mRNA expression levels were also analyzed in microarray data of a series of B-cell lymphoma cases including 9 GCB-DLBCL, 9 ABC-DLBCL, 15 mucosa associated lymphoid tissue (MALT) lymphoma, 15 follicular lymphoma (FL) and 12 SMZL specimens (G), as well as analyzed by Q-RT-PCR in biopsies of 119 DLBCL cases and compared between patients with high (above cohort median) and low BCL6 expression or mortality score (H). LITAF mRNA expression assessed by Q-RT-PCR increased in the BCL6 expressing GCB-DLBCL cell line OCI-Ly1 after BCL6 silencing with a siRNA (I, upper panel) or BCL6 inhibition with the BPI peptide (I, lower panel). Q-RT-PCR data was normalized to the corresponding controls as 2-ΔΔCt. Boxplots of three independent experiments are shown. LITAF increase was also confirmed at protein level by Western Blot (J). (K) ChIP-Seq enrichment around LITAF exon 1 (small black square under the enrichment histograms) corresponding to two different anti-BCL6 antibodies (N3 and D8). The location of the primer pairs used for Q-ChIP experiments are depicted at the bottom. The enrichment of the sequence between the second primer pair was confirmed by Q-ChIP (L), using an anti-actin antibody as negative control. Data are displayed as mean±SD of 2 independent experiments. This sequence (Seq2) was cloned in a luciferase expression plasmid controlled by the SV40 promoter and showed a repressive effect abolished by the mutation of the putative binding site for BCL6 located in this sequence (Seq2 Mut) in Dual Luciferase assays (M). Mean±SEM data of 4 independent luciferase experiments in OCI-ly1 cells are shown. ** means statistical significance with p value below 0.01, n.s., non-significant differences (p> 0.05). CB, centroblasts; MC, memory B cells; Naïve, naïve B cells.
