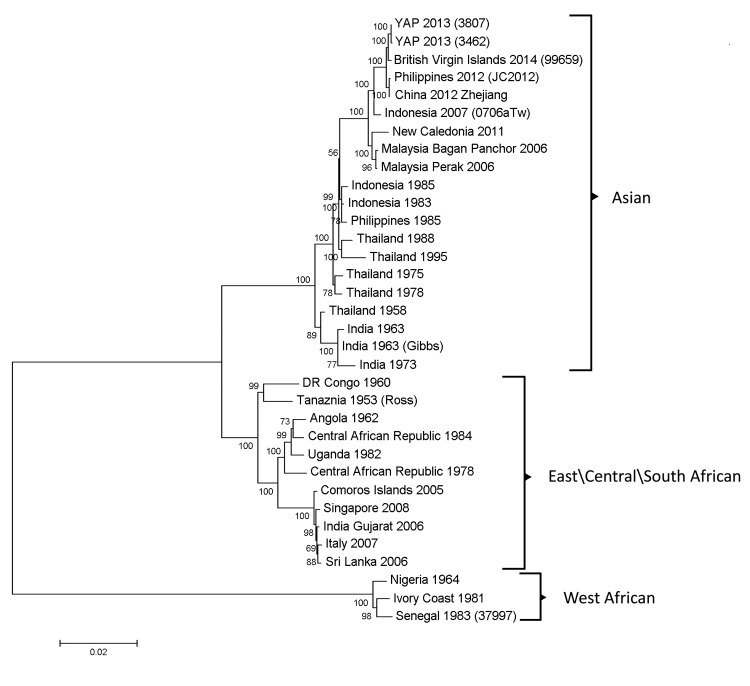
Figure.
Phylogenetic tree derived by neighbor-joining methods (1,000 bootstrap replications) using complete genome sequences of chikungunya viruses obtained from GenBank. Scale bar represents the number of nucleotide substitutions per site. Genotypes are indicated on the right. GenBank accession numbers for viruses used for construction of the tree follow: Yap 2013–3807 (KJ451622), Yap 2013–3462 (KJ451623), British Virgin Islands-99659 (KJ451624), Philippines 2012-JC2012 (KC488650), China 2012 Zhejiang (KF318729), Indonesia 2007–0706aTw (FJ807897), New Caledonia 2011 (HE806461), Malaysia 2006 Bagan Panchor (EU703759), Malaysia 2006 Perak (EU703760), Indonesia 1985 (HM045797), Indonesia 1983 (HM045791), Philippines 1985 (HM045790), Thailand 1988 (HM045789), Thailand 1995 (HM045796),Thailand 1975 (HM045814),Thailand 1978 (HM045808),Thailand 1958 (HM045810), India 1963 (HM045803), India 1963 Gibbs (HM045813), India 1973 (HM045788), DR Congo 1960 (HM045809), Tanzania 1953 Ross (AF490259), Angola 1962 (HM045823), Central African Republic 1984 (HM045784), Uganda 1982 (HM045812),Central African Republic 1978 (HM045822), Comoros 2005 (HQ456251), Singapore 2008 (FJ445510), India 2006 Gujarat (JF274082), Italy 2007 (EU244823), Sri Lanka 2006 (GU189061), Nigeria 1964 (HM045786), Ivory Coast 1981 (HM045818), and Senegal 1983–37997 (AY726732).