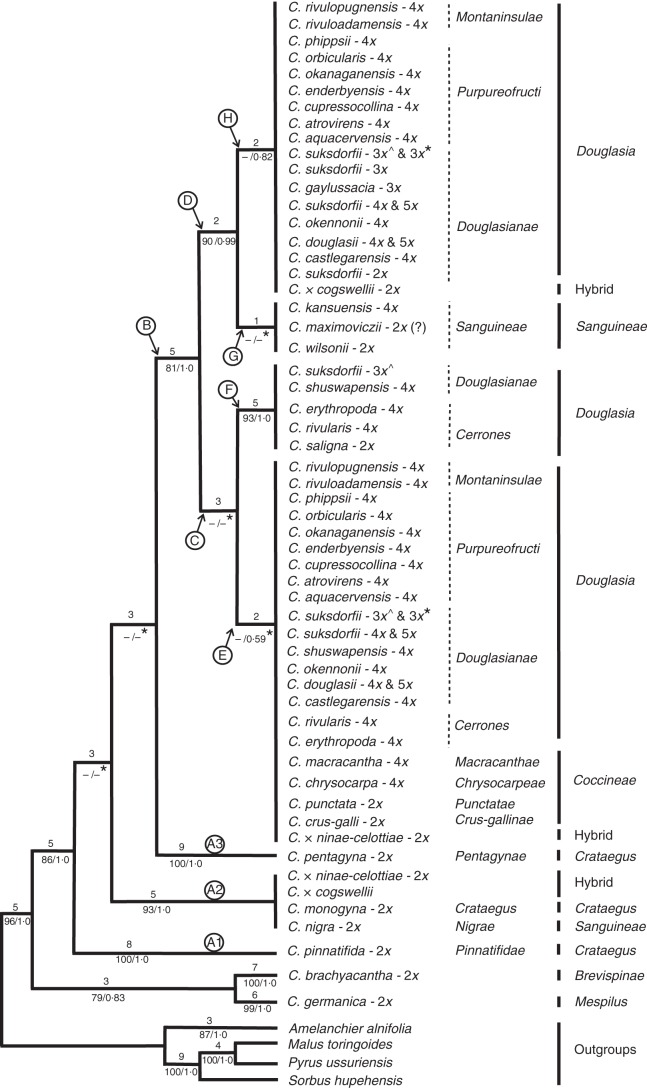
Fig. 3.
One of the most-parsimonious trees, randomly selected from >100 000 trees, and obtained from analysis of the data matrix of ITS2 sequences (299 OTUs). Tree length = 458, consistency index (CI) = 0·55, rescaled consistency index (RC) = 0·5, homoplasy index (HI) = 0·45 and retention index (RI) = 0·92. Branch lengths (ACCTRAN optimization) are indicated above branches and bootstrap percentages and posterior probabilities below branches (BS/PP). – indicates branches with BS <50 and collapsed in the 50 % majority-rule consensus tree of Bayesian inference. The branches in each main clade were collapsed into polytomies to simplify the tree visually. Ploidies are those reported in Talent and Dickinson (2005), Lo et al. (2007, 2013) or J. Coughlan and N. Talent (unpubl. data). ‘C. suksdorfii - 3x*’ indicates the accession sampled from Vancouver Island (JC385; Supplementary Data Table S1), and ‘C. suksdorfii - 3x̂’ represents the accession sampled from Haida Gwaii (2010-42; Table S1). An asterisk (*) after PP values below branches indicates the branches collapsed in the strict consensus of >100 000 most-parsimonious trees.