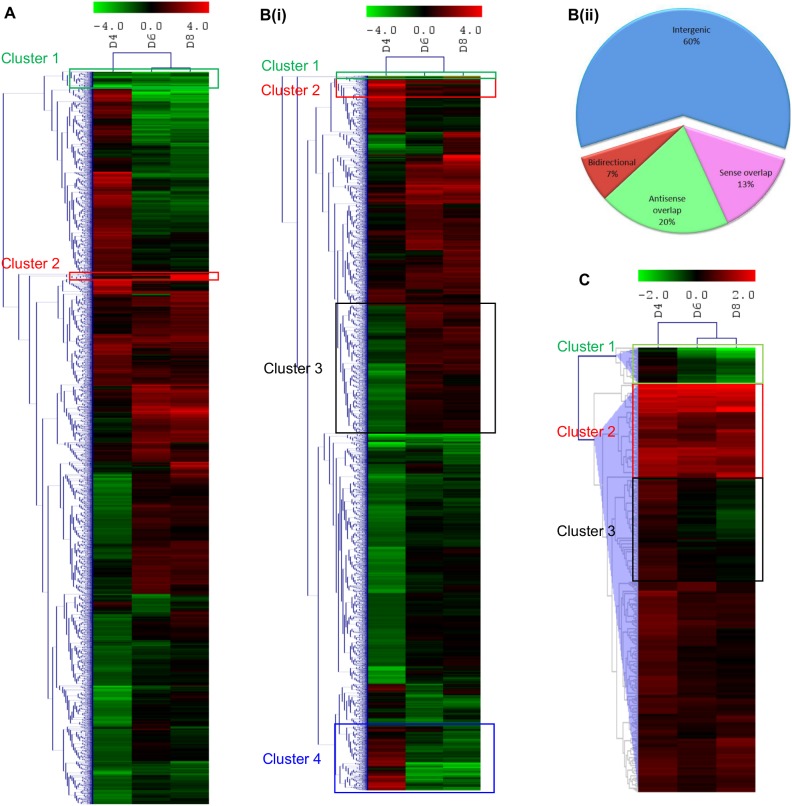
Figure 2. Transcriptome of maturing neurons.
(A) Hierarchical clustering analyses of mRNAs in maturing cortical neurons. Clusters of highly down- and up-regulated mRNAs were identified. Bi) lncRNAs in maturing neurons. Clusters of highly down- and up-regulated lncRNAs with their orientation to the genome or proximal gene loci were identified. (Bii) Subgroup analyses of altered lncRNAs in relation to their nearby coding genes. (C) miRNAs in maturing cortical neurons. Clusters of highly down- and up-regulated miRNAs were identified. Total RNA from 4 separate experiments (n = 4) carried out in triplicates were pooled for each time points. The microarray analyses were carried out for each time point on the pooled RNA. The average signal intensities were 369.03, 501.12, 429.58 and 455.03 for the lncRNA and mRNA microarray for Day 2, Day 4, Day 6 and Day 8 respectively. For miRNA microarray, the average signal intensities were 1673.43, 1783.01, 1385.10 and 1698.38 for Day 2, Day 4, Day 6 and Day 8 respectively. Hierarchical clusters were constructed out using average linkage and Euclidean distance as the similarity measure. Green rectangle indicates downregulation and red, upregulation.