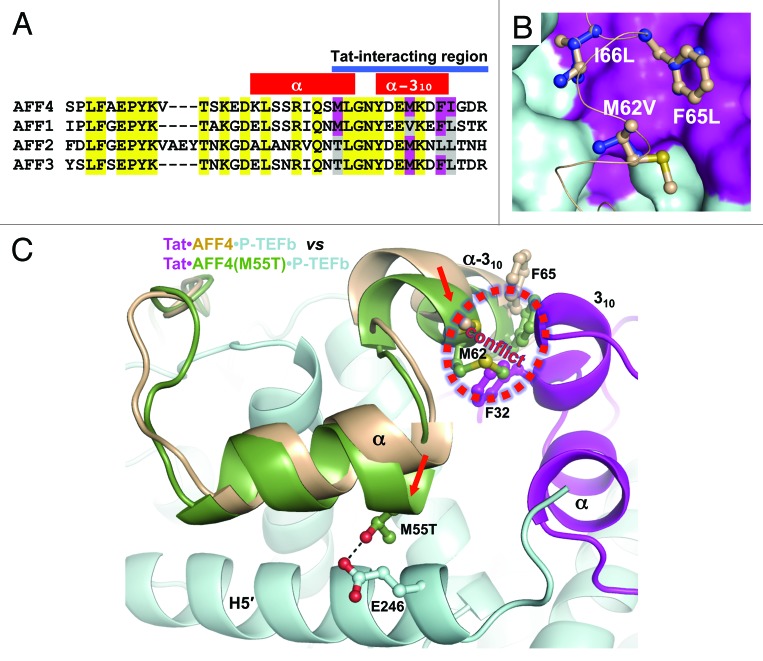
Figure 5. Analysis of Tat binding by AF4 family members. (A) Amino acid sequence alignment of AFF4 CBS with AFF1, AFF2 and AFF4 sequences. Conserved sequences are marked in yellow. Tat-interacting hydrophobic sequences of AFF4 that are not conserved are marked in magenta. Corresponding residues in other AF4 family members are marked in magenta if they are the same as in AFF4 or gray if they are different than in AFF4. (B) Modeling of Met62Val, Phe65Leu and Ile66Leu substitutions. Original residues are in light orange and substituted residues are in blue. (C) Possible effect of Met55Thr substitution in AFF2 and AFF3. The Cyclin T1 domains of AFF4•P-TEFb and Tat•AFF4•P-TEFb were superimposed. Then the AFF4 domain from AFF4•P-TEFb with a modeled Met55Thr substitution was displayed together with Tat•AFF4•P-TEFb. Met55Thr shifts AFF4 α-helix toward the Cyclin T1 helix H5′ and forms a hydrogen bond with Glu246 of Cyclin T1. As result, the AFF2 or AFF3 conformation will be closer to the AFF4 conformation in AFF4•P-TEFb structure. This would cause a conflict between the α-310-helix of AFF2 or AFF3 and 310-helix of Tat. Red arrows indicate the directions of shifts caused by Met55Thr substitution.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.