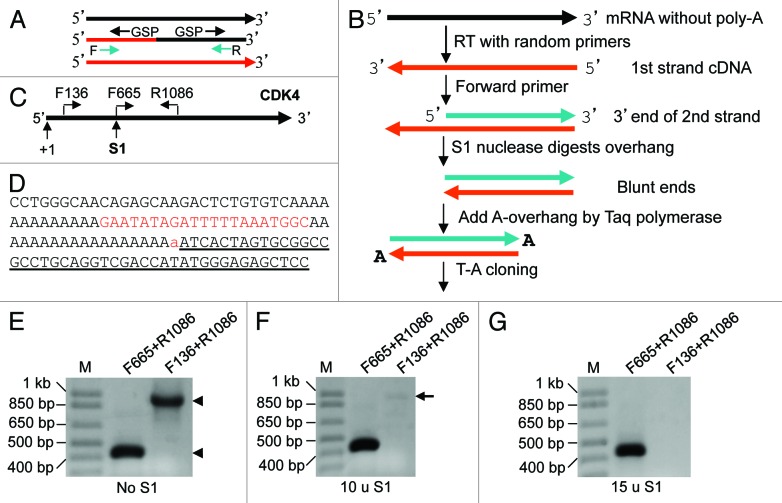
Figure 1. Cloning RNA 3′ end. (A) Two hurdles for cloning the 5′ or 3′ end, and for PCR amplification, of chimeric cDNA: (1) Gene-specific primer (GSP) used in RACE amplifies the 5′ or 3′ end of not only the chimeric cDNA but also the cDNA of a parent gene (black or red line). (2) Forward (F) or reverse (R) primer primes not only the chimeric cDNA but also the cDNA of a parent gene, making the first several cycles of PCR less efficient. (B) Our strategy for cloning RNA 3′ end: After RT with random hexamers, a forward primer of the gene of interest and Taq are used to synthesize the 3′ part of the second cDNA strand. S1 is added to cut the 3′-overhang of the first strand. The cDNA blunt ends are then appended with a dA by Taq, followed by T-A cloning. (C) Illustration of the locations of primers and the S1 cutting site on the CDK4 mRNA. (D) Part of the 3′ sequence obtained, in which the lowercase “a” is added by Taq and the underlined sequence belongs to the T-A vector. The sequence matches completely to the CDK4 mRNA. Note that there is an internal poly-A sequence 21 nt upstream of the authentic poly-A tail. (E–G) Both pairs of primers (F665+R1086 and F136+R1086) designed to amplify the 5′ and the middle regions of the mRNA, respectively, can amplify the RT product (cDNA) of RNA from HeLa cells without S1 digestion (arrowheads). F655+R1086 can still yield a band from the cDNA digested with 10 or 15 units of S1, while the F136+R1086 yielded only a very faint band (arrow) when 10 units were used and no band when 15 units were used.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.