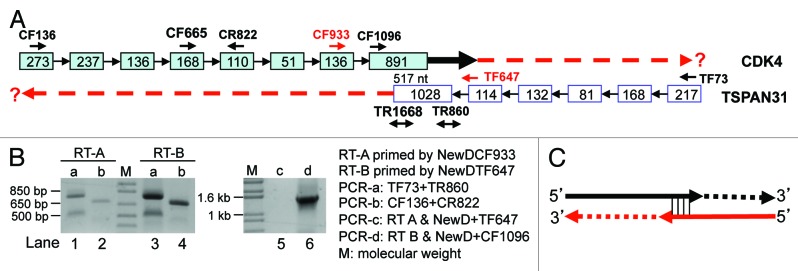
Figure 5. RT with linker-containing GSP to detect CDK4 (NM_000075.3) and TSPAN31 (NM_005981.3). (A) Illustration of the CDK4/TSPAN31 relationship according to the NCBI, with the locations of CDK4 forward (CF) or reverse (CR) primers and TSPAN31 forward (TF) or reverse (TR) primers indicated. Boxes represent exons with their length indicated as the number of nt. Note that the two mRNAs overlap at their last 517 nt. (B) RT of unDNased RNA from Hela cells primed by the NewDCF933 should specifically convert the TSPAN31 mRNA to cDNA (RT-A), whereas RT primed by the NewDTF647 should specifically convert the CDK4 mRNA to cDNA (RT-B), if the mRNAs reach the regions of these primers. These two RT products were used as the template in PCR with either the TF73+TR860 (PCR-a, lanes 1 and 3) or the CF136+CR822 (PCR-b, lanes 2 and 4) as the primer pair or in PCR with the primer pair of NewD+TF647 (PCR-c, lane 5) or NewD+CR1096 (PCR-d, lane 6). The results in lanes 4 and 6 together with sequence data suggest that some CDK4 transcripts reach the TF647 region (thick black arrow in A). (C) When sense or antisense RNA has an unprotected 3′ end overlapping with the other, the overlapping sequence may serve as a primer in RT to extend its 3′ end with its antisense as the template. The extension may also occur in the ensuing PCR, as depicted in Figure 6A. Because the mRNA does not really have this extended part (dashed lines), the corresponding PCR product, such as the bands in lanes 1, 2 and 3 in (B), is a wrong-template artifact.
