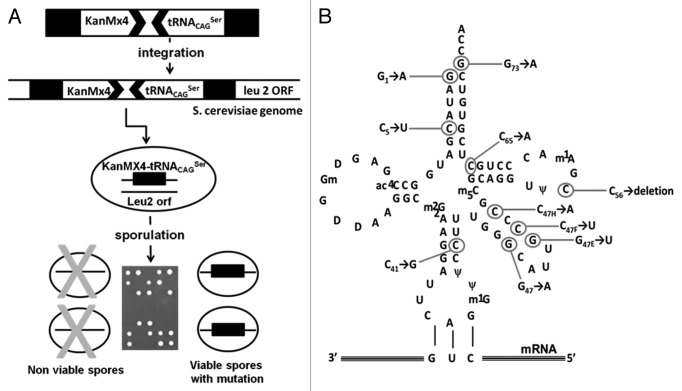
Figure 5. In vivo forced evolution methodology used to identify mutations that inactivate the C. albicans tRNACAGSer. (A) In vivo forced evolution was used to screen for mutations that affect the C. albicans tRNACAGSer stability and identity. This method was based on PCR amplification of the DNA cassette containing the tRNACAGSer fused to the KanMx4 gene and subsequent integration by homologous recombination into the LEU2 locus of diploid yeast cells. Yeast containing the tRNACAGSer gene integrated in the genome were sporulated and tetrades were dissected using a micromanipulator. The tRNACAGSer gene was amplified from viable spores by PCR and the amplified DNA was sequenced. (B) The in vivo forced evolution strategy using the tRNACAGSer gene expressed in yeast cells identified 10 different mutations. Most mutations localized in the acceptor-stem and variable-arm.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.