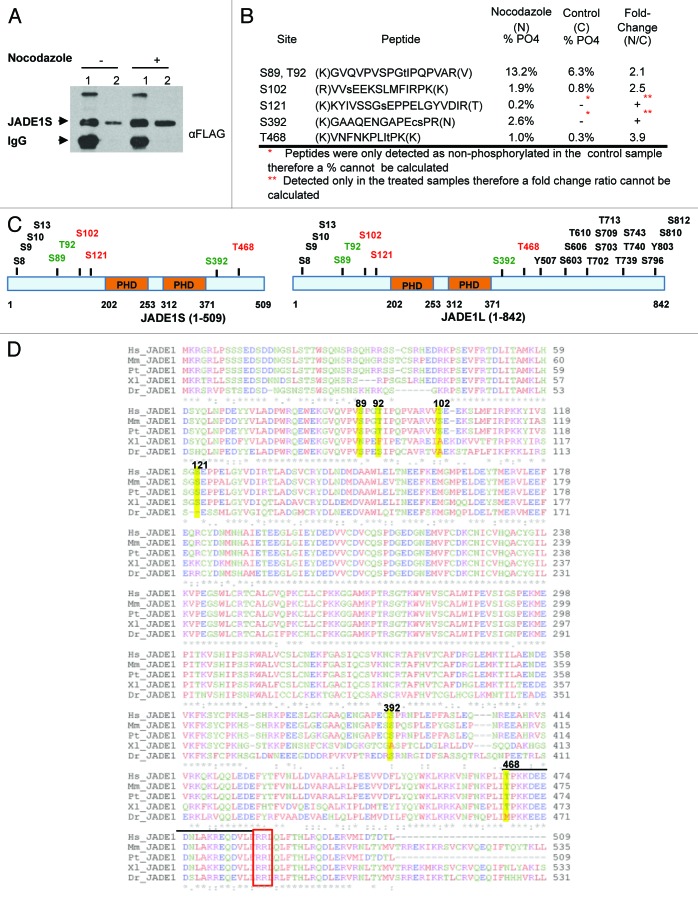
Figure 9. Mass spectrometry analysis of JADE1S during nocodazole-induced G2/M cell growth arrest identified 6 amino acid residues that are phosphorylated. H1299 cells were co-transfected with FLAG-JADE1S and HA-HBO1, and subsequently treated with nocodazole or vehicle. (A) JADE1S-HBO1 complex affinity purification. Sample IP, 7.5% SDS-PAGE. Lane 1; JADE1S protein bound to A/G agarose beads, lane 2; JADE1S protein eluted from beads. (B) Phosphorylation sites were identified in JADE1S by nano-LC/MS/MS. Using peak areas of the unphosphorylated peptide and the phosphorylated peptide the percentage of phosphorylation was calculated, and the fold change ratio was determined. Note that phosphorylation of S121 and S392 was not detected in the control sample (red asterisk). (C) In silico analysis of JADE1 phosphorylation sites (black and green; http://www.phosphosite.org) and sites identified through MS analysis (red and green). (D) Sequence alignment of JADE1 protein in various species showing the conservation of the identified phosphorylation sites (ClustalW2). Hs, Homo sapiens; Mm, Mus musculus; Pt, Pan troglodytes; Dr, Danio rerio; Xl, Xenopus laevis. Yellow highlights indicate the identified amino acid residues. Note that lined region contains CDK binding site sequence (S/T-P-X-R/K) and consensus cyclin binding motif RRL (red box).

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.