Figure 1. AGT structures.
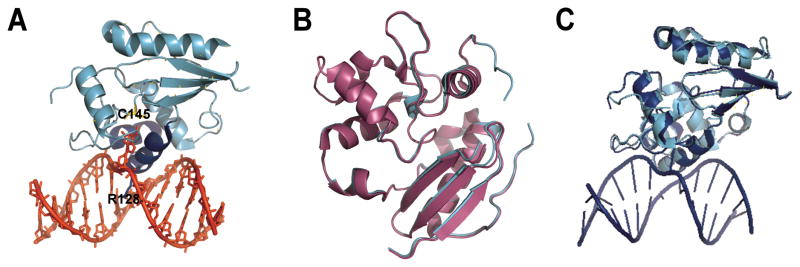
(A) AGT (cyan and blue) binds to the minor groove of the DNA double helix (shown in red) via a helix-loop-helix motif (blue), leading to flipping of the alkylated base into the active site cleft of the protein where it is transferred onto a cysteine (C145). An arginine finger (R128) stabilizes the void left by the flipped base. (Coordinates from 1T38.pdb) (B) Overlay of the alkylated (magenta; 1EH6.pdb) and the apo (cyan; 1EH7.pdb) forms of AGT. Alkylation destabilizes a short alpha helix near the active site. (C) Overlay of the AGT monomer bound to a DNA lesion (blue; 1T38.pdb) with the unbound protein (cyan, 1EH6.pdb). DNA binding does not introduce significant structural changes in AGT (RMSD 0.6 Å, determined with DaliLite, EMBL-EBI).
