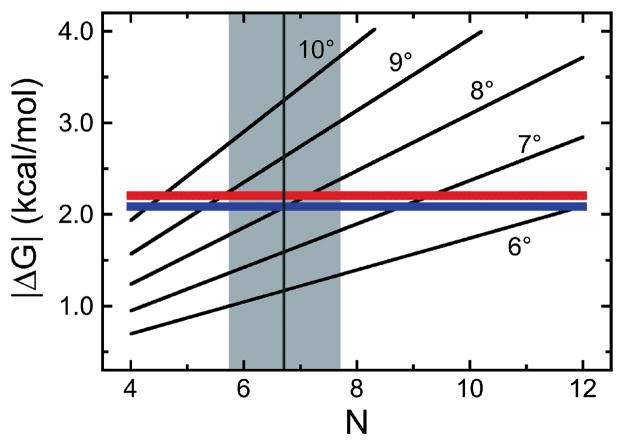
Figure 11. Limiting energetics in AGT cluster formation.
[7] The quadratic model (Eq. 5) was used to calculate the cumulative ΔG(DNA twist) as functions of proteins/cluster (N) for DNA twist angles of 6–10 degrees/protein. The red and blue lines give |ΔG(cooperative)| (Eq. 6) for the addition of a single protein molecule to complexes formed on linear pUC19 DNA and the 1,000 bp fragment, respectively. The intersections of these functions indicate where ΔG(DNA twist) = −ΔG(cooperative). A vertical grey line (grey zone between minimum and maximum estimates of cluster length) is plotted for a cluster length of 6.7 proteins, the mean of all length estimates for [AGT] ≥ 6μM from AFM experiments with the 1,000 bp DNA.