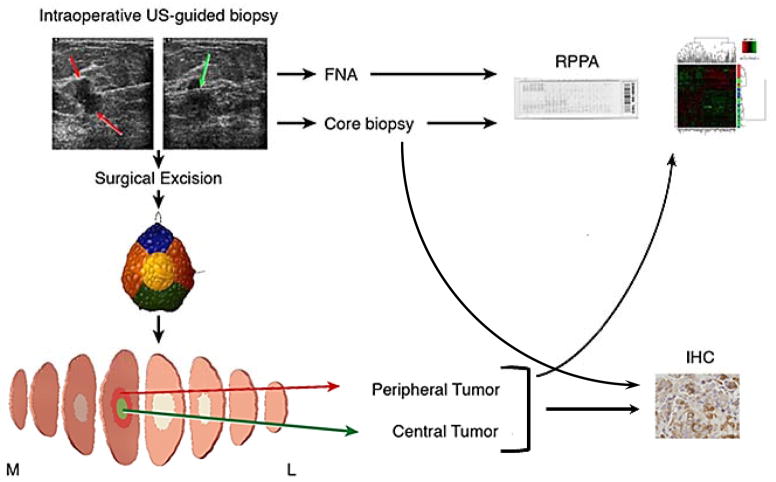
Figure 1.
Study Schema. Patients underwent intraoperative breast biopsies with fine needle aspirate (4–8 passes) and core needle biopsy (3–4 passes). The core needle biopsy with the greatest tumor cellularity was used for RPPA while the other core needle biopsies were analyzed by IHC. After surgical excision the specimen underwent routine intraoperative assessment including color-coded specimen margin inking, and sectioning and specimen mammogram as needed. Central and peripheral tumor specimen was collected for research to capture tumor heterogeneity. FNA, core needle biopsy and peripheral and central surgical resection samples underwent proteomic analysis with reverse phase protein arrays (RPPA). Core needle biopsy and both peripheral and central surgical resection samples underwent IHC for PI3K pathway markers.