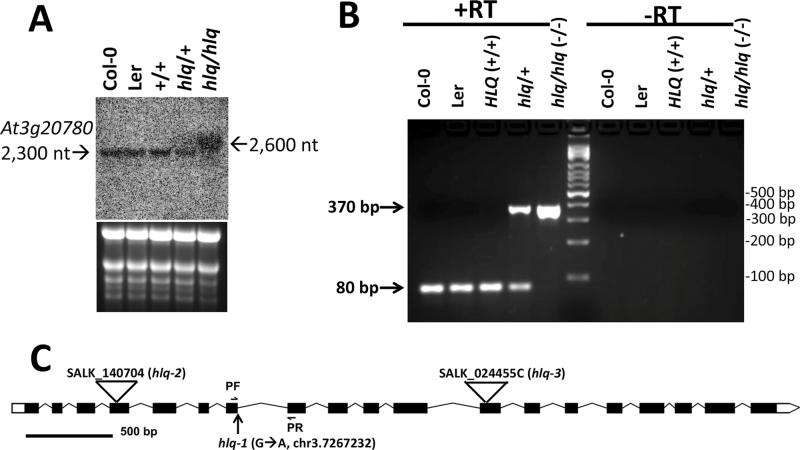
Fig. 4.
Characterization of the At3g20780 mRNA in the hlq mutant demonstrates aberrant intron 7 splicing. (A) RNA gel blot of At3g20780 shows an aberrant mRNA in hlq homozygotes. A 20 μg aliquot of total RNA from seedlings of wild types Col-0, Ler, and HLQ+/+ (abi2-1 parental background), hlq/+ heterozygotes, and the homozygous hlq/hlq mutant were probed with a 1.5 kbp fragment of the At3g20780 cDNA; the lower panel shows equal loadings by ethidium bromide staining of the denaturing agarose gel before blotting. Band sizes of the wild-type and hlq At3g20780 mRNAs are labelled on the left and right sides, respectively. (B) Agarose gel of RT–PCR amplicons from cDNA prepared from total RNAs of control (Col-0, Ler, and HLQ/HLQ), hlq/+ heterozygotes, and hlq/hlq homozygotes (–/–) shows retention of intron 7 (355bp predicted) in hlq genotypes, and absence of intron 7 (~80bp amplicon, 55bp predicted) in controls. ‘+RT’, with reverse transcriptase; ‘–RT’ reverse transcription step omitted from the amplification protocol. (C) Cartoon showing the exon–intron–untranslated region (UTR) structure of At3g20780, the position of primers (PF, PR; horizontal arrows) used for PCR in B, the position of the hlq-1 mutation (vertical arrow) at the intron 7 splice donor site, and positions of T-DNA insertions (triangles) in lines SALK_024455C (hlq-3) and SALK_140704 (hlq-2).