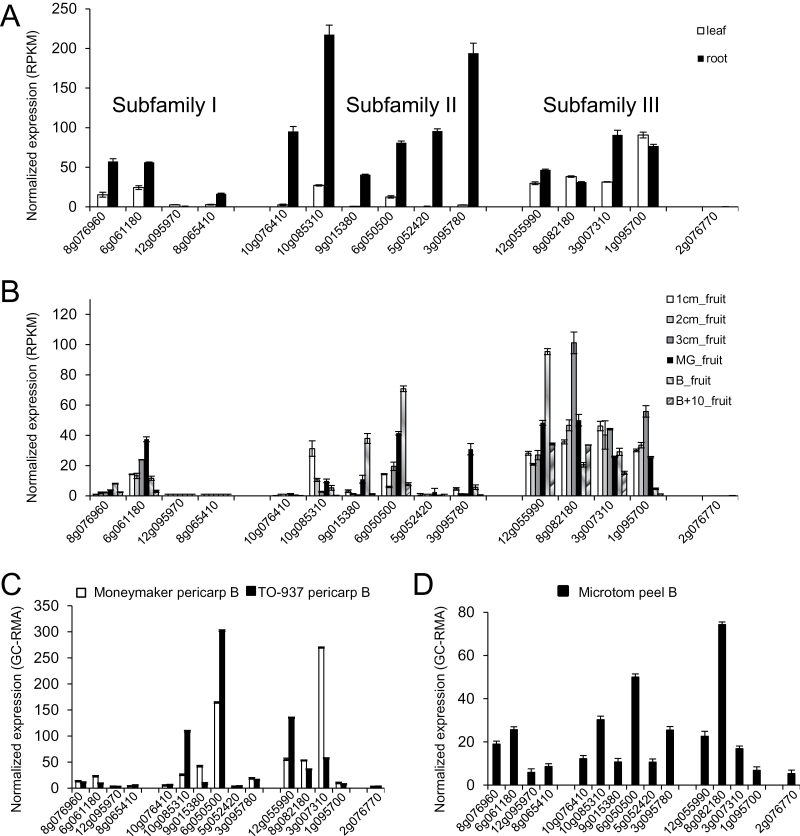
Fig. 2.
Relative gene expression of tomato ABA receptors in leaf, root, and fruit was determined by RNA-Seq and microarray analysis. (A, B) The transcriptome of the inbred tomato cultivar Heinz 1706 was analysed using Illumina RNA-Seq technology (Tomato genome Consortium, 2012). Data show gene transcription of tomato receptors grouped in three subfamilies and the ungrouped 2g076770 in leaf and root (A) and during fruit development and ripening (B). Significant expression of 2g076770 and 12g095970 was not detected in these tissues. RPKM, reads per kilobase of exon model per million mapped reads; MG, mature green stage; B, breaker stage. (C, D) Relative RNA abundance based on GC-RMA values (GC content–robust multiarray analysis) obtained from the Affymetrix exon tomato microarray (EUTOM3) hybridized with fruit pericarp RNA of Moneymaker and TO-937 (C) and fruit epidermis RNA of the Microtom background (D) at the breaker stage.