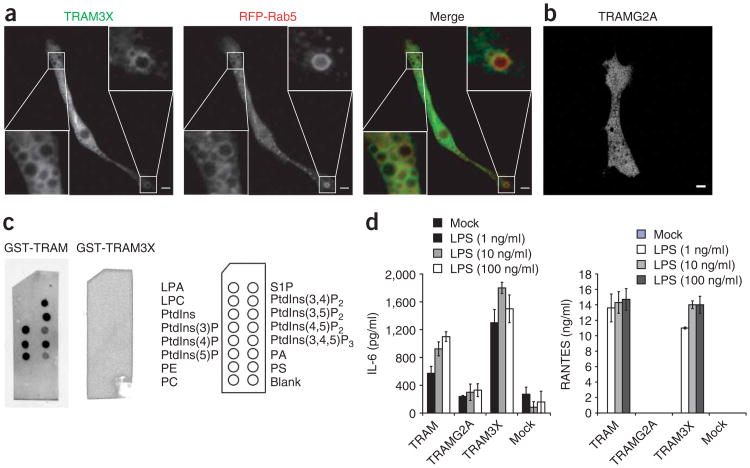
Figure 4.
TLR4 can induce the production of type I interferon from early endosomes. (a,b) Fluorescence microscopy of macrophages transfected with plasmids encoding a GFP-tagged TRAM construct with substitutions in the polybasic domain (TRAM3X) and RFP-Rab5 (a) or with a myristoylation-deficient TRAM mutant (TRAMG2A; b). Outlined areas are enlarged in the bottom left and top right corners (a). Scale bars, 5 μm. Data are representative of at least three independent experiments with over 500 cells per condition, in which over 95% of the cells had similar staining patterns. (c) ‘PIP strips’ of various lipids (identified at right) overlaid with GST-tagged wild-type TRAM (left) or TRAM with substitutions in the polybasic domain (middle); lipid binding was identified by standard protein-protein immunoblot techniques. LPA, lysophosphatidic acid; LPC, lysophosphatidylcholine; PE, phosphatidylethanolamine; PC, phosphatidylcholine; S1P, sphingosine 1-phosphate; PA, phosphatidic acid; PS, phosphatidylserine. Data are representative of two independent experiments. (d) ELISA of IL-6 and RANTES in TRAM-deficient macrophages transduced with retroviral vectors encoding GFP-tagged TRAM mutants (horizontal axes) and challenged with 1, 10 or 100 ng/ml (key) of LPS. The transduction efficiency of each resulting macrophage population was determined to be 30–40% by flow cytometry and fluorescence microscopy. Data are representative of three independent experiments.