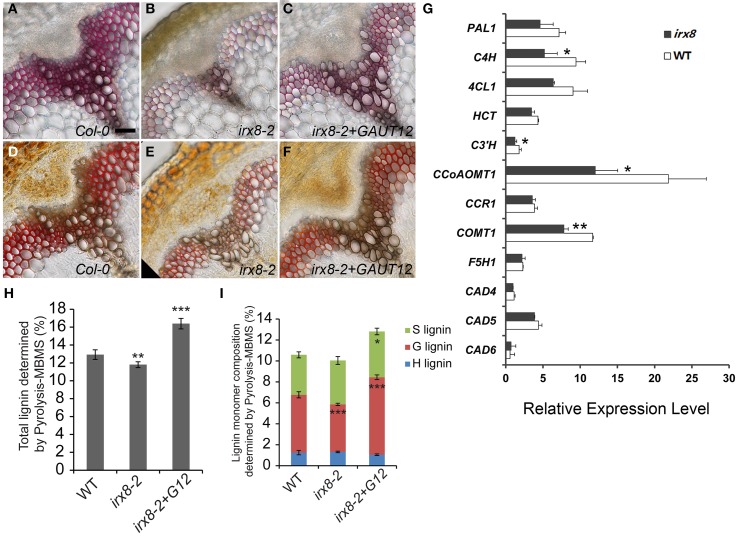
Figure 4.
Measurement of lignin and lignin biosynthetic gene expression in the irx8 mutant. Phloroglucinol-HCl staining of free-hand basal stem cross-sections of WT (A), irx8-2 (B), and irx8-2+GAUT12 (C). Mäule staining of free-hand basal stem cross-sections of WT (D), irx8-2 (E), and irx8-2+GAUT12 (F). Bar = 50 μm for A–F. (G) Expression analyses in Arabidopsis WT and irx8 lower stems of lignin biosynthetic genes using Real-Time PCR. Genes labeled with asterisks have significantly lower expression in irx8-5 compared to WT (*p < 0.05; **p < 0.01). The relative expression level of each gene was normalized using Actin2 as the reference gene and the expression of C3′H in wild-type basal stem was set to 1. Values are mean ± standard deviation (n = 3). Lower-stem refers to the lower half of the inflorescence. PAL, phenylalanine ammonia lyase; C4H, trans-cinnamate 4-hydroxylase; 4CL, 4-coumarate: CoA ligase; HCT, hydroxycinnamoyl-CoA: shikimate/quinate hydroxycinnamoyltransferase; C3′H, 4-coumaroyl shikimate 3′-hydroxylase; CCoAOMT1, caffeoyl-CoA 3-O-methyltransferase; CCR, cinnamoyl-CoA reductase; F5H, ferulate 5-hydroxylase; COMT, caffeic acid/5-hydroxyferulic acid O-methyltransferase; CAD, cinnamyl alcohol dehydrogenase; PER/LAC, peroxidases/laccases. (H) Total lignin (%) determined by pyrolysis molecular beam mass spectrometry (pyMBMS) of stem alcohol insoluble residues. (I) Lignin monomer composition determined by pyMBMS. Lignin content percentage (%) was corrected by equivalence to Klason lignin. The irx8-2 and irx8-2+GAUT12 values were compared to WT as determined by ANOVA and post-hoc t-tests with Bonferroni correction (*p < 0.05; **p < 0.01; ***p < 0.001; n = 6) for (H,I).