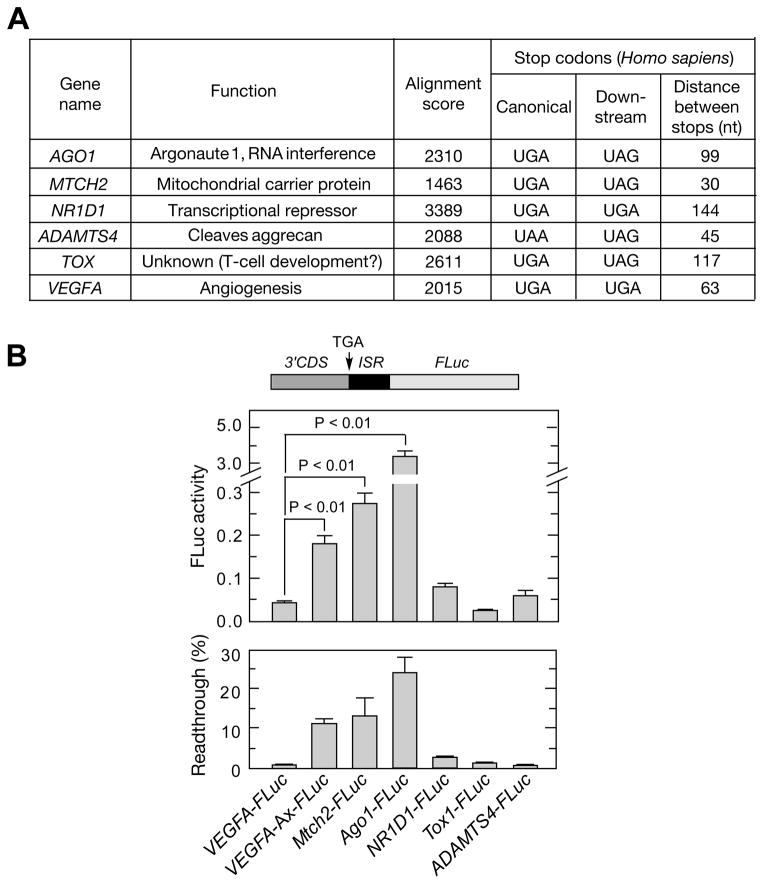
Figure 7. Genome-wide Analysis to Identify Targets of Translational Readthrough.
(A) Properties of candidate genes selected for experimental validation. Candidates were selected using a bioinformatic screening protocol (Figure S6).
(B) Validation of translational readthrough targets. Chimeric plasmids containing about 700 nt of the 3′-terminus of human coding sequences upstream of the inter-stop codon region and in-frame with FLuc were transfected into HEK293 cells. Relative FLuc activities normalized to FLuc mRNA were determined by qRT-PCR (top). Readthrough efficiency is expressed as percent of FLuc activity of normal translation controls in which canonical UGA stop codon is replaced by GCA (bottom).
See also Figure S6.