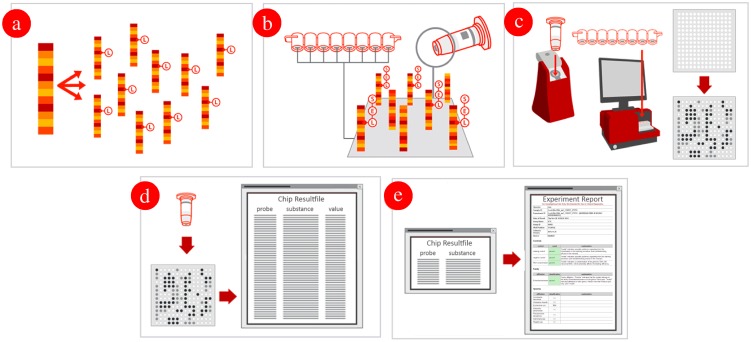
Figure 1. Linear multiplex DNA amplification, labeling and hybridization with the ArrayStrips.
(a) Linear Multiplex Amplification starting from clonal RNA-free genomic DNA. Extracted DNA is internally labeled with biotin (Label [L]) and amplified in a linear multiplex PCR reaction; (b) Hybridization: the internally biotin labeled, single-stranded DNA product hybridizes specifically under stringent conditions to the corresponding probes. The resulting duplex is detected using a horse-radish peroxidase (Enzyme [E]) – streptavidin conjugate, which causes the dye to precipitate ([S]). (c) Detection: the ArrayMate Reader (or ArrayTube Reader ATR 03) enables the visualization and subsequently automated analysis of the array image. The presence of a dark precipitated spot indicates successful hybridization; (d) Analysis: the assay specific software analysis script coming with the ArrayMate Reader (or ArrayTube Reader ATR 03), measures the signal intensity of each probe and determines which genes/alleles are present in the sample by means of an assay specific algorithm. (e) Genotype analysis: a software plugin coming with the ArrayMate Reader (or ArrayTube Reader ATR 03) analyzed the raw data automatically, finally a report is provided on the detected carbapenemases genes.